Future-proofing Cybersecurity in Drug Discovery
The pharmaceutical and biotech sectors suffer more data security breaches than any other industry, with 53% resulting from malicious activity. To protect against potential ...
News
The next Design a Molecule competition begins on March 15th, 2012 and we’re so excited about the next target, we’re giving you a little sneak peek. Like the last competition, the theme is anti-malarial, but this time we’ve chosen to focus on on a broad spectrum protein target relevant to antibacterials, antifungals and various plasmodium-like human parasites.
The enoyl acyl carrier protein reductase (ENR/FABL/FABI) is involved in fatty acid biosynthesis in bacteria, specifically in the liver stage of plasmodium falciparum life-cycle (PfENR) and more generally in that of Toxoplasmodium Gondi (TgENR).
Triclosan which is a broad spectrum anti-bacterial used in various applications, such as antibacterial soaps and toothpaste, is well established as an inhibitor of ENR’s. More recently Triclosan has been shown to inhibit the human parasitic forms of plasmodium (PfENR and TgENR) showing some efficacy in mice – although there is controversy over the precise mechanism and involvement of PfENR as the primary target. A number of X-ray structures are available of ENRs and triclosan analogues (below).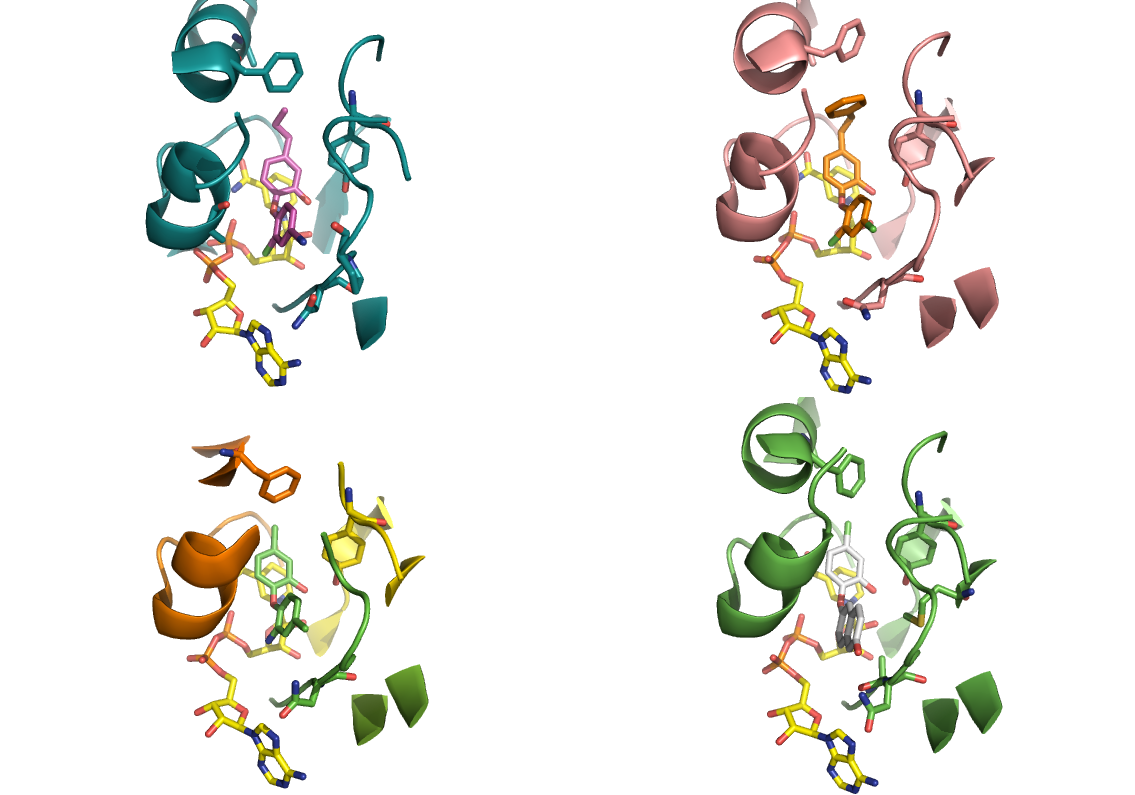
Conversion of Triclosan into a drug has a number of associated challenges:
1. Efficacy – less active sub-optimal at PfENR relative to bacteria
2. Toxicity – poly chlorination is an issue
3. Solubility – too lipophilic logP X TpSA Y – although has to pass through multiple membranes to access target
4. Resistance – an issue in bacteria
Recent examples of optimisation of Triclosan for Plasmodium ENR have appeared in the literature – some of which may also have potential against resistant bacteria, as in the figure below.
We have created an artificial 3D ligand construct by combining some of the desirable ligand complex examples from the PDB in order to address poly-chlorination issues. It should be noted that this is simply a ‘virtual ligand template’, representing accessible ligand –protein interface within PfENR, which we hypothesise to be useful for scoring potential designs against – and is not a known active.
The challenge in this competition is therefore to design a ligand that addresses the issues above as much as possible. The winning entry will have the highest score against this construct whilst having the lowest 2D similarity. The winner will also be judged on synthetic tractability and the extent to which the design addresses the issues described above (1-4).
The competition runs March 15th – April 20th and we’ll give you more detailed instructions along with the 3D molecule file when registration opens. If you’re new to Design a Molecule, here’s how it works:
1. Register for the competition
2. We’ll give you a target molecule and free licenses to FieldAlign and FieldView
3. Design the very best molecule
4. Submit the molecule to us
5. Win an iPad2 (Well, only if you really designed the very best molecule in the bunch)
Feel free to contact us with questions about our target selection, the competition or using Cresset software for molecule Design.
Update March 15: The competition is now open, register here and see the instructions here.