Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Today the UK’s most powerful GPU-based supercomputer, ‘Emerald’, will enter into service alongside the ‘Iridis 3’ system at the Science and Technology Facilities Council’s Rutherford Appleton Laboratory (RAL) in Oxfordshire, UK. These two High Performance Computing systems will give businesses and academics unprecedented access to super-fast processing capability.
Cresset is collaborating with the high performance computing group at the University of Bristol, UK to implement new GPU based algorithms within the core of our field technology. The following poster will be presented at today’s meeting. For further details of our project with the University of Bristol refer to our Fields at Warp Speed blog post.
Mark Mackey†, Simon McIntosh-Smithµ, Simon Krige†, Rob Scoffin†
†Cresset Biomolecular Discovery Ltd, BioPark, Broadwater Rd, Welwyn Garden City, Herts, AL7 3AX, UK
µDepartment of Computer Science, University of Bristol, Woodland Road, Clifton, BS8 1UB, UK
It has long been known that small molecule drugs are recognized by and bind to proteins on the basis of their 3D electronic and shape properties, yet the drug discovery cycle has traditional described and protected 2D structures.
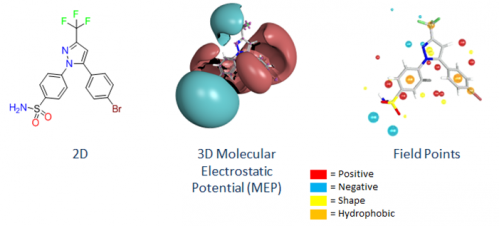
Cresset is using field point descriptions of molecules to close the gap between chemistry and biology, bringing the features that are recognized by proteins to the desktop of our customers.
Field Points are a condensed representation of electrostatic, hydrophobic and shape properties (protein’s view).
Molecular Field Extrema
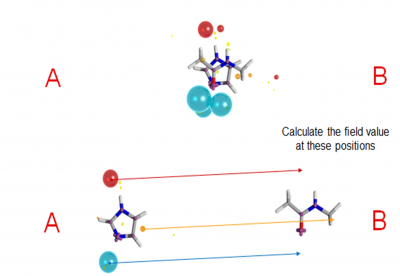
Given an alignment:
– For a given field point on molecule A, calculate what the field value is at the corresponding point in molecule B. The score of the field point is the product of its size and B’s field value.
– Repeat for all field points on A and calculate the sum of scores
– Repeat for the field points on B sampling the field of A, and normalise to a similarity
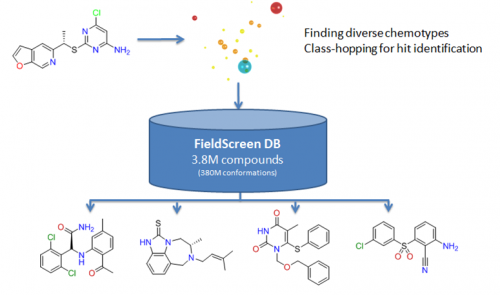
Current Situation
Large database of molecules (~5million)
– Compute time: 2-5s per molecule on a single CPU core
– Full screening takes ~35hours on 200 CPUs
– Full screening costs ~$500 on CPU
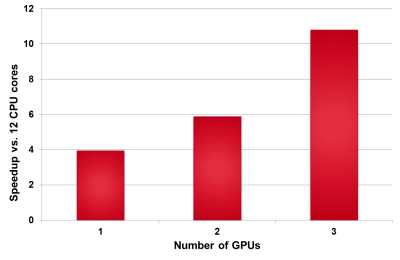
Using GPUs and the Emerald Cluster
We have run the prototype FieldScreen GPU port on Emerald nodes. Speedup results are relative to the serial code running on 12 Intel i7 CPU cores.
– Using OpenCL: currently ~40x faster for a GPU vs a CPU.
– Full screening:
~$20 on GPU (25 times cheaper than CPU).
~30min using all Emerald GPUs!
The Emerald Cluster is giving us the opportunity to screen large virtual libraries of compounds (> 100m compounds) in very little time. The speed and cost advantages of GPUs have made it our technology of choice.
Funded by a TSB Knowledge Transfer Partnership.