Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
A new version of Forge is available now. This version brings many new and improved features and is recommended for all users. Of particular note are:
Forge is Cresset’s computational workbench to understand SAR and design. Free evaluations are available by simply registering for a demo or contacting us for pricing and licensing options.
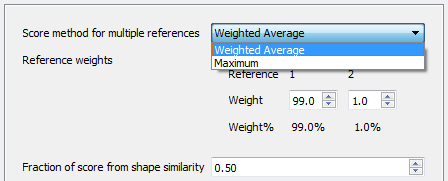
Weighting multiple reference molecules
Using multiple reference molecules in Forge is a good way to add extra information to your alignment and 3D-QSAR experiments. Incorporating references that bind to different parts of the active site of the protein or incorporate diverse functionality significantly improves the alignment of diverse active molecules. However, the alignment step has always assumed that the reference molecules are equally important.
In this release we have introduced the ability to make reference molecules more or less important by modifying the proportion of the score that is derived from that reference. Now the handling of multiple reference molecules becomes very flexible, giving you the option to control the weighting scheme, for example by using the maximum score against any reference or by using a weighted average of scores.
This capability will help with SAR interpretations that involve many chemotypes and also with design experiments where you are trying to build a set of interactions that exist in one series into another.
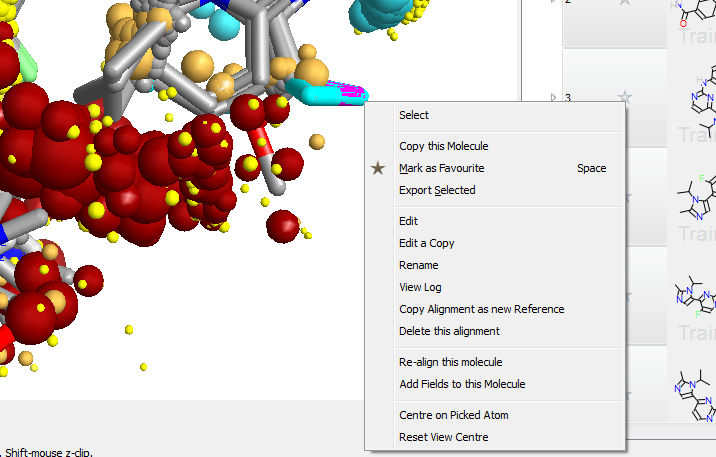
A new context sensitive menu
A new context sensitive menu on the 3D window makes examining and modifying alignments simpler than ever. Right clicking on an atom gives you options to select, copy or edit the selected molecule.
When several molecules are displayed at once, this simple new capability becomes a powerful way of selecting molecules with interesting alignments, of modifying interesting compounds and of repairing mis-aligned molecules.
Enhanced molecule viewing capabilities
This release of Forge brings enhanced molecule viewing capabilities. We have taken all the capabilities of Cresset’s legacy FieldView product and added them into Forge. This gives you the ability to load, sketch and view molecules, including their field patterns, as well as to load and view Blaze results files.
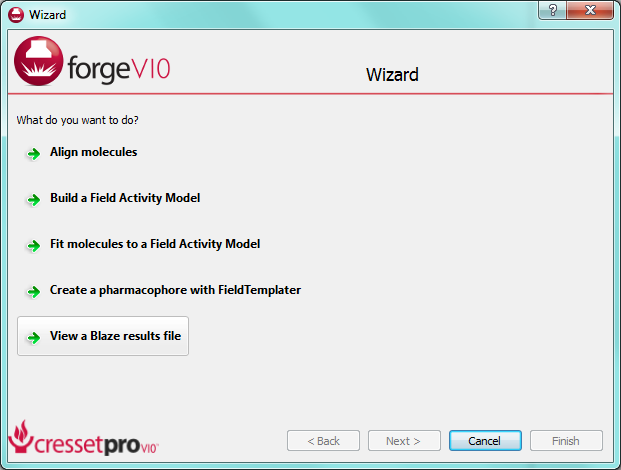
Loading a Blaze result file in ForgeV10.1 is now a smooth process. Reference, protein and database molecules are auto-recognized and displayed appropriately. To make life easier still, we’ve added a wizard entry to give rapid access to the analysis of Blaze results (see screenshot, below).
A simple protein import process
The simple protein import process that we released in torchV10 has been introduced in this release of Forge. Aimed at the rapid and simple processing of pdb files into reference ligands and protein excluded volumes, the pdb import has been an instant hit with Torch users. Using a hierarchical, sortable view of the protein enables the loading specific protein chains, removal of all or just “hot” waters molecules, and rapid identification of the bound ligand. This short Forge video shows the process.
Forge is Cresset’s computational workbench to understand SAR and design. Free evaluations are available by simply registering for a demo or contacting us for pricing and licensing options.