Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Cresset’s field based fragment replacement software ‘Spark’ was designed from the outset to tie its results to synthetic chemistry. This led to options to limit the nature of new bonds that are formed and to sorting our fragment libraries on frequency of occurrence. These options, combined with the superior electrostatics of Cresset’s XED force field for scoring the fragment replacement, gives novel, synthetically accessible molecules in many projects.
In the latest release of Spark we have expanded the synthetic accessibility concept yet further by the introduction of fragments from specific reagent pools. This enables rapid searching of all the possible R-groups that we could include at a specific position for those that best fit our multi-parameter optimization problem. If you are using the database generator tool then the search space becomes tightly linked to your available chemistry – you create a database for the specific reagents that you have in stock, enabling a search of the chemistry space that you can access today.
In this case study we used Spark’s predecessor (FieldStere) in a virtual thought experiment exploring the possibility of using the tool for growing a fragment bound to P38 kinase. We used two inhibitors that had been co-crystallized and deposited into the PDB as 3K3I and 3ROC as our starting points (Figure 1) and tried to grow the smaller inhibitor to be ‘like’ the larger, DFG-in inhibitor.
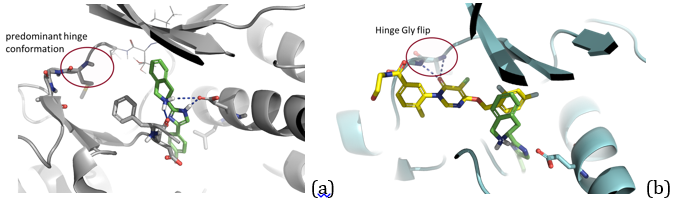
Initially the experiment was run starting from the 3K3I derived inhibitor as the fixed component and a dummy atom provided in the ortho position of the tetrahydro isoquinoline (TIC) moiety as the site for fragment replacement. The output field similarity score was weighted 80:20 between these two references derived from the two inhibitors – 3ROC : 3K3I. A protein excluded volume was not used.
In autumn 2011 we were pleasantly surprised and excited to see that the results of this exercise were interestingly very close to a structure deposited by Pfizer (Figure 2).
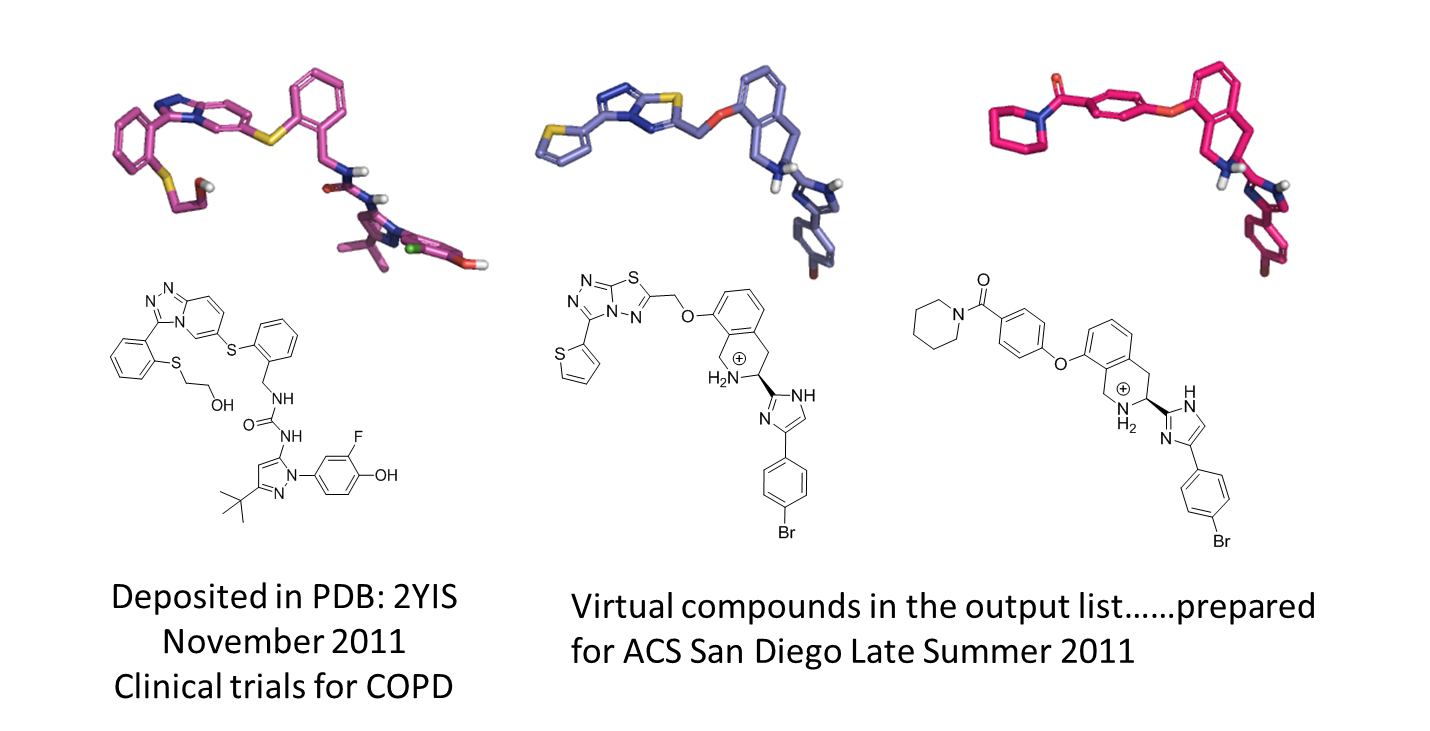
We decided to reproduce this experiment again but this time using the protein from 2YIS as an excluded volume and using the new reagent pool databases in Spark.
Assuming that the same chemistry that was used to construct the Pfizer compound can be applied to the TIC example the searching a database of commercially available thiols should provide readily synthesizable results.
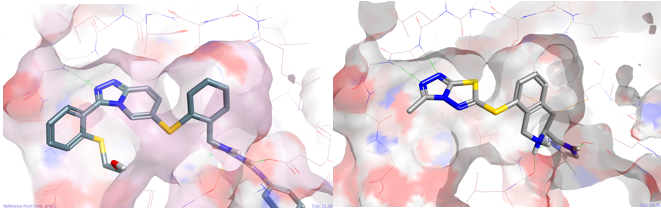
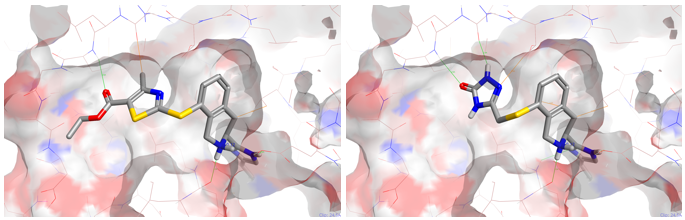
The third result in the list is shown in Figure 3. This new design shows a beautiful fit into the P38 protein in keeping with the initial thought experiment. Two further examples are shown below (Figure 4) again from the commercial pool which would also be applicable using the same chemical transformation.
Interesting output was also obtained using different reagent pools e.g. amines and alcohols which would involve the use of different chemical transformations but possibly via the same or a different starting chemical TIC intermediate.
The new Spark reagent databases enable rapid profiling of the accessible chemistry space around a specific core. Using the Spark database generator enables generation of reagent databases that are linked tightly to your immediately available moves.