Future-proofing Cybersecurity in Drug Discovery
The pharmaceutical and biotech sectors suffer more data security breaches than any other industry, with 53% resulting from malicious activity. To protect against potential ...
News
 Most of Cresset’s applications require good conformations for molecules in order to produce excellent results. This could be for pharmacophore elucidation (in FieldTemplater module of Forge), virtual screening (in Blaze), searching databases of fragments for new bioisosteres (in Spark) or relating known or newly designed molecules in Forge and Torch. Wherever we need them, we want the best possible ensemble of conformations in the fastest time. Inevitably most approaches (including ours) involve a trade-off between completeness, diversity and speed.
Most of Cresset’s applications require good conformations for molecules in order to produce excellent results. This could be for pharmacophore elucidation (in FieldTemplater module of Forge), virtual screening (in Blaze), searching databases of fragments for new bioisosteres (in Spark) or relating known or newly designed molecules in Forge and Torch. Wherever we need them, we want the best possible ensemble of conformations in the fastest time. Inevitably most approaches (including ours) involve a trade-off between completeness, diversity and speed.
In our GUI tools, we’ve generally exposed three recommended conformation hunt pre-sets:
We are constantly trying to hone our algorithm to improve the results we get and understand where our current compromise lies. Our original pre-sets worked well but given that computers are significantly faster now than when we originally designed the Accurate setting, it seemed worth taking another look to see if we could improve the results further by spending a few more CPU cycles.
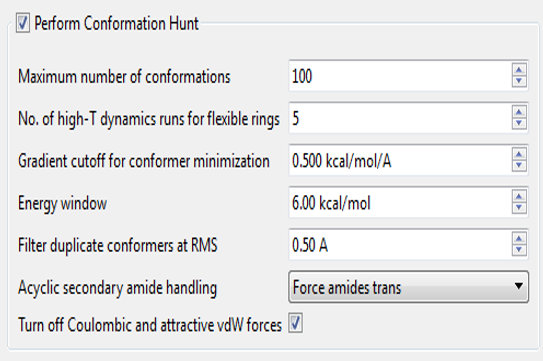 The two parameters we focused on were the energy window and the minimization gradient cut-off. The energy window refers to how high a conformation’s energy has to be relative to the lowest-energy conformation (an approximation to the global minimum) before we throw it away as being “too strained”. Reports in the literature suggest that values of at least 3-5kcal/mol1,2 to more than 25kcal/mol3 are needed if you are to keep the bioactive conformation. However, it is quite hard to define what energy function you should use to calculate the “strain energy” of binding. In vacuo energies are clearly inappropriate. However, water-phase energies are also somewhat inappropriate, as ‘protein phase’ has a much lower dielectric than water – it depends how much of the full thermodynamic cycle of binding you want to assign as part of the conformer energy.
The two parameters we focused on were the energy window and the minimization gradient cut-off. The energy window refers to how high a conformation’s energy has to be relative to the lowest-energy conformation (an approximation to the global minimum) before we throw it away as being “too strained”. Reports in the literature suggest that values of at least 3-5kcal/mol1,2 to more than 25kcal/mol3 are needed if you are to keep the bioactive conformation. However, it is quite hard to define what energy function you should use to calculate the “strain energy” of binding. In vacuo energies are clearly inappropriate. However, water-phase energies are also somewhat inappropriate, as ‘protein phase’ has a much lower dielectric than water – it depends how much of the full thermodynamic cycle of binding you want to assign as part of the conformer energy.
Given the need for speed and the difficulty of computing ‘correct’ conformation energies (as well as the difficulty in describing what ‘correct’ means), we fall back on an extremely simple pseudo-solvation model: all long-range electrostatics are ignored, as are all attractive vdW forces. This model is very cheap computationally and provides quite good conformations. The conformer “energies” calculated using this model are more a measure of total internal strain, rather than being true conformation energies and tend to have significantly less spread.
In the past, we have used 6 kcal/mol as our energy window. This is a fairly wide window, but was necessitated by the fact that in the default Normal conditions for our conformation enumerator we don’t minimize the conformations very hard (minimization gradient cut-off ~ 0.5 kcal/A). This is partly for speed purposes, but also because bioactive conformations are not necessarily local minima in whatever force field you want to use.1 Using a loose minimization cut-off and a large energy window gives good success rates when tested against a standard “can you find the bioactive conformer” test set, but there is a cost. You occasionally get unrealistic/unlikely conformations, especially of saturated rings – axial substituents, twist-boats and so on. Our use of a precomputed ring conformation library reduces this problem but does not eliminate it.
In this study, we found that minimizing the conformations harder (gradient cut-off ~0.1 kcal/A2) allows us to reduce the energy window to 3 kcal/mol without losing our ability to find the bioactive conformers (see table). In the process, you ensure that conformers with locally large strain energies such as twist-boats are filtered out, improving the overall quality of the population and reducing the number of conformers generated. The price, however is time: the new method is 4-5x slower than the old one. We felt that this was a trade-off that was worth taking but if you disagree then it is easy to create your own settings that correspond with the old Accurate (contact our support team if you would like help with this).
| Settings | Normal | Old Accurate | New Accurate | |
|---|---|---|---|---|
| Max Confs kept | 100 | 200 | 200 | |
| Energy Window | 6 | 6 | 3 | |
| Minimization RMSL | 0.5 | 0.5 | 0.1 | |
| % bioactive confs within (Å RMSD) |
0.25 | 8 % | 10 % | 10 % |
| 0.50 | 43 % | 49 % | 49 % | |
| 0.75 | 66 % | 68 % | 70 % | |
| 1.0 | 79 % | 82 % | 82 % | |
| 1.5 | 92 % | 94 % | 92 % | |
| 2.0 | 98% | 99% | 99% | |
| Relative time | 10 | 19 | 97 | |
| Avg nos. of confs | 88 | 152 | 124 |
The new settings are in the last release (V10.3) of Forge, Torch and Spark. So, if you’ve been wondering why the ‘Accurate but slow’ settings seem slower than before, that is why. The good part is that even though it’s more ‘slow’, it’s also more ‘Accurate’ now!