Future-proofing Cybersecurity in Drug Discovery
The pharmaceutical and biotech sectors suffer more data security breaches than any other industry, with 53% resulting from malicious activity. To protect against potential ...
News
A common concept in medicinal chemistry is the bioisostere. The exact definition of a bioisostere is rather fuzzy, but the Wikipedia entry is as good as any: ‘bioisosteres are chemical substituents or groups with similar physical or chemical properties which produce broadly similar biological properties to another chemical compound.’ Bioisosteric replacements come in two general classes: core replacements, where the centre of a molecule is changed, creating a new chemical series; and leaf replacements where a replacement is sought for part of the molecule on the periphery, keeping it in the same series. Core replacements can get you out of a difficult ADMET or IP situation or can be a useful stepping stone to developing a backup series. Leaf replacements are more common as part of the day-to-day work of lead optimization – having discovered a substituent that works, the obvious thing to do is to try more things like it!
There are many software products available to search for bioisosteric replacements, both commercial and freely-available. The proliferation of methods is largely because searching for bioisosteres looks so easy. Look at the piece you want to replace, search a database for something largely the same size and geometry, and present the results. Simple, right? However, if you delve into it, the whole process is much more complex than it appears …
The first issue for a bioisostere replacement method involves deciding whether a fragment is physically the right size – are the connection points in the right place and at the right angle? Again this seems a trivially easy question to address until we consider that this is a 3D question. You need to consider the 3D geometry of the fragment – what conformation is it in? We chose to use a mixed approach to this: we provide both fragments derived from the CSD where the conformation comes directly from the small molecule crystal structure and large databases of fragments where we provide a small distribution of low energy conformations. As a result, you can just search experimental small molecule crystal conformations if you wish, but you can augment that search with a much larger and richer conformation database if you so desire.
The second criterion is pharmacophoric: a bioisosteric replacement must have ‘broadly similar biological properties’ and hence its interactions with the protein must be similar to the original molecule. Again, there’s a subtlety here. If I’m looking for a replacement for a triazolothiazole, then when I’m presented with a candidate fragment the obvious thing to do is ask “how similar is it to triazolothiazole?” This approach is flawed because the properties of the candidate fragment (and the initial triazolothiazole!) depend on their environment i.e. the rest of the molecule. This is especially true if you are interested in molecular electrostatics, as the electrostatic potential of a molecule is a global property that cannot generally be piecewise decomposed. That is why Spark performs all scoring in product space, not in fragment space (Figure 1). However, even if you are using a more crude method such as shape or pharmacophore similarity, treating the fragment in isolation can be very misleading.
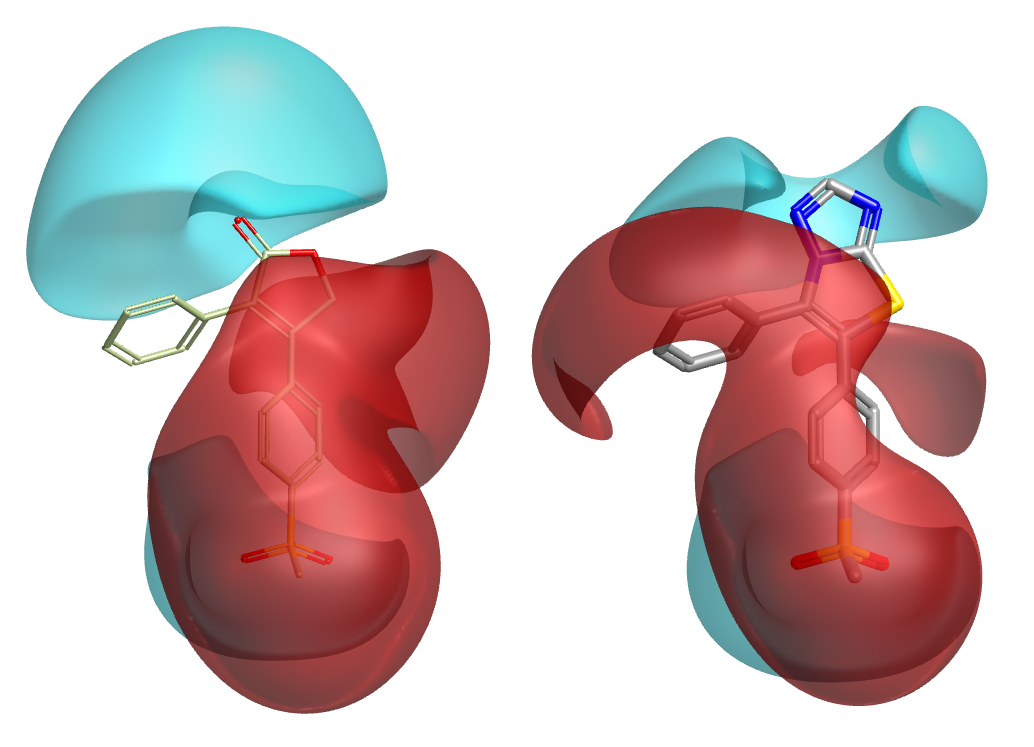
Working in product space brings advantages in steric considerations as well as electronic. Even though a particular fragment might have a beautiful geometric fit, and match the original core very well in terms of shape/electrostatics/color/pharmacophores/whatever, you still need to assess whether the fragment is compatible with the conformation of the original molecule. That assessment cannot be done in fragment space! In Spark we handle this in two ways. Firstly, all product molecules are minimized prior to scoring. If, due to steric or electronic effects, the candidate fragment just is not compatible with the desired conformation of the original molecule, then this minimization will distort the invariant parts and lead to a very low similarity score. Fragments which cause steric clashes or lone pair-lone pair repulsions are thus automatically filtered out (Figure 2). Secondly, we follow this up with an explicit assessment of strain energy around the newly-formed bonds, so the user can immediately see whether there are any causes for concern.

Another thing that can be drastically affected by the environment is the hybridization and charge state of the fragment (and the rest of the molecule). The main culprit here is nitrogen. Is an amine basic or not? Is the nitrogen pyramidal or trigonal planar? Both these questions depend strongly on what the nitrogen is attached to and hence you cannot assess the suitability of a fragment without reference to the chemical environment that you are going to place it in. These questions turn out to be surprisingly difficult to handle in a completely robust way: there is a large amount of code in Spark which assesses the local chemical environment around the newly-formed bonds and determines whether any hybridization or formal charge changes are required. It turns out you cannot just recharge the product molecule, as in some cases the user may have assigned particular charge states to parts of the molecule (based on experimental knowledge of the charge state when bound to the protein, for example) and you don’t want to undo that.
The final complication to performing the scoring properly (i.e. in product space) is accounting for the available degrees of freedom. If you are doing a core replacement, then the positon of the core is generally completely determined by the attachment points. However, if you are doing a leaf replacement, then the question arises as to what rotamer to choose around the attachment bond. The only solution is to perform a limited conformation search around that bond and score each conformer, which has the potential to get very slow although there are some computational tricks you can do to reduce the search space. It’s not just leaf groups that have to be searched: replacing the centre of a molecule might require a rotation scan if there are two attachment points which happen to be collinear.
So, our nice and simple bioisosterism calculation has become rather more complicated. Rather than just see if a fragment fits into the hole in the original molecule, we need to merge it in properly, assess any hybridization or formal charge state changes that are required, minimize the resulting molecule, perform a rotation scan around any underspecified degrees of freedom, recompute the electrostatic potentials, align to the original molecule and calculate the steric and electrostatic similarities. Only then can we decide whether the fragment is any good or not! Luckily, Spark does all of that for you, so as a user you can concentrate on the results, rather than on the complicated calculations required to produce those results.
Try it for yourself – download a free evaluation of Spark.