Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
The new release of Spark comes with new and updated fragment and reagent databases. These are designed to give you the widest sources of inspiration for your projects, whilst also enabling a close link between Spark’s suggestions and the chemistry that is available to you.
The latest Spark databases include over 3.5 Million fragments that are used to find novel bioisosteres for your project. These come from two distinct sources – ZINC and ChEMBL. In each case the molecules from the entire source collection are fragmented and the frequency with which any fragment has appeared is noted. We then sort the fragments according to frequency and label them according to the number of bonds that were broken to disconnect the fragment from its original molecule.
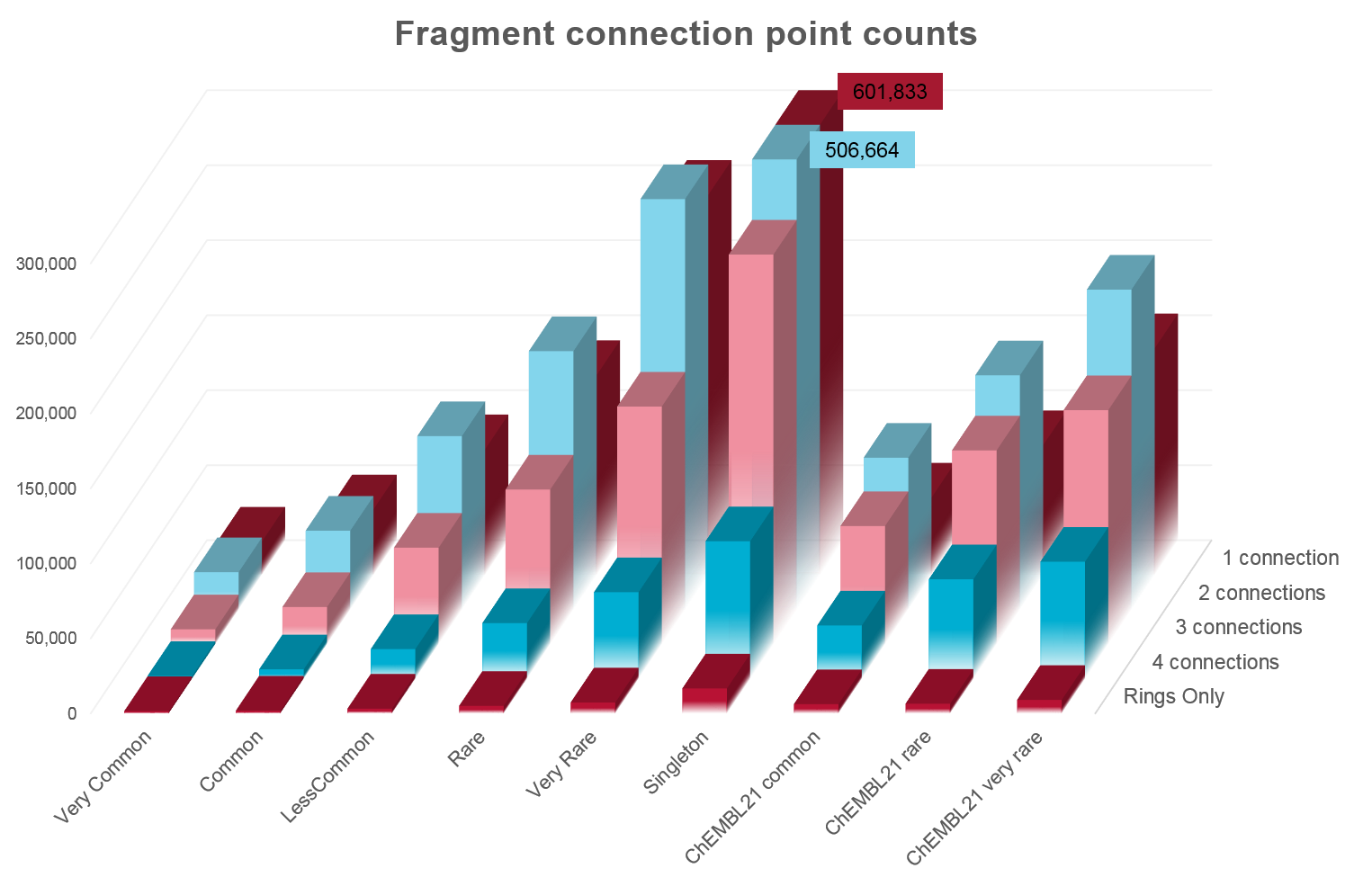
Analysis of the numbers of fragments in common between the ZINC and ChEMBL databases shows surprising complementarity.
| Number of fragments (to nearest 1000) |
Very Common | Common | Less Common | Rare | Very Rare | Singleton |
|---|---|---|---|---|---|---|
| ChEMBL21 common | 41,000 | 41,000 | 43,000 | 26,000 | 18,000 | 15,000 |
| ChEMBL21 rare | 7,000 | 24,000 | 44,000 | 46,000 | 45,000 | 34,000 |
| ChEMBL21 very rare | 3,000 | 11,000 | 26,000 | 34,000 | 42,000 | 70,000 |
Unsurprisingly, there is significant overlap in the most common fragments from each database. However, once you get to the rarer fragments it is apparent that ZINC and ChEMBL occupy quite distinct parts of chemical space, with the majority of “rare” fragments being unique to each database.
In this release we have completely replaced the source of our reagent fragments. We are delighted to be working with eMolecules to provide you with over 500,000 reagents that are easy to order with known availability. The new eMolecules based reagent databases use an enhanced set of rules to more closely relate the Spark results to the chemistry that you want to use on your molecules.
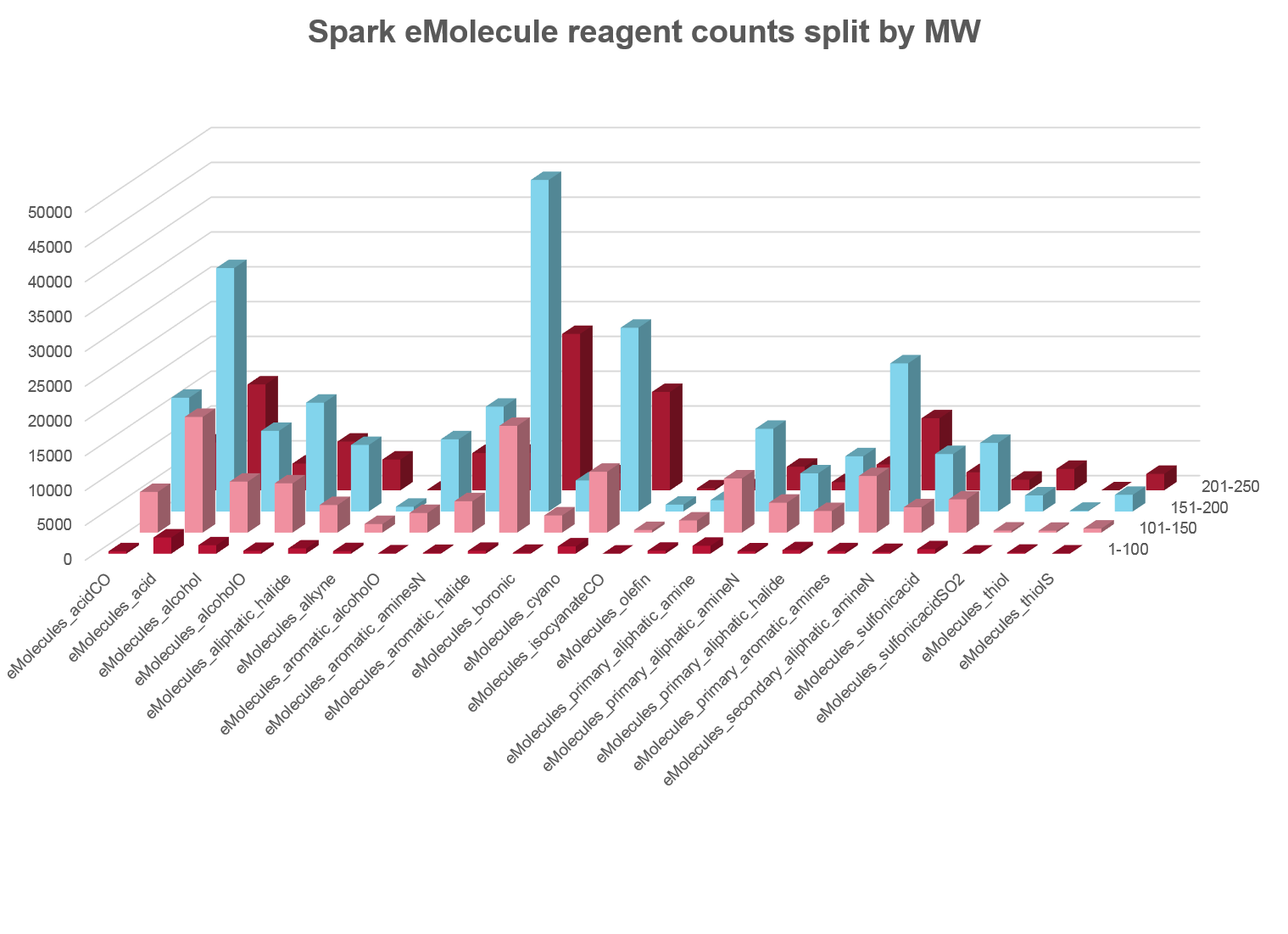
Each fragment in the eMolecules database is linked back to both the eMolecules ID for the source reagent and its availability. Running a Spark search on these databases thus allows you to very simply move from the Spark experiment to ordering the reagents you require to turn your Spark results into reality.
This release of databases for Spark increase the number of fragments and the improve the availability of reagents. When combined with the existing VEHICLe derived database, the CSD derived database and databases from your corporate collections generated with Spark’s database generator we believe that Spark will find a even better range bioisosteres for your project.
To update to the latest databases or to take a look at how Spark can impact your project please contact us.