Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Activity Atlas1 is a component of Forge2, Cresset’s powerful workbench for ligand design and SAR analysis. Activity Atlas models summarize the SAR for a series into a visual 3D model that informs design decisions and helps prioritize molecules for synthesis. In this case study, Activity Atlas’ activity cliff summary maps were used to analyze the SAR of a small series of Nav1.7 sodium channel antagonists. The objective was to investigate and understand the electrostatic, hydrophobic and shape features underlying receptor activity in a case where crystallographic information is not available for the target protein.
Structural information is becoming commonly available even for those targets, such as GPCRs and ion channels, which until recently were considered difficult to crystallize.
For novel targets and new chemical series, however, X-ray data may still be difficult to obtain. Quite frequently the information available to project chemists during the early stages of a discovery project can be so scarce to severely hamper the applicability of traditional structure- and ligand- based computational approaches such as docking and pharmacophore modeling.
A method capable of quickly identifying and deciphering the most relevant features underlying protein-ligand interaction, starting from a very limited amount of Structure-Activity Relationship (SAR) data and no structural information, would be of invaluable help during the early stages of drug discovery projects.
We introduced Activity Atlas, a probabilistic method of analyzing the SAR of a set of aligned compounds as a function of their electrostatic, hydrophobic and shape
properties. The method uses a Bayesian approach to take a global view of the data in a qualitative manner. Results are displayed using Forge visualization capabilities to gain a better understanding of the features which underlie the SAR of your set of compounds.
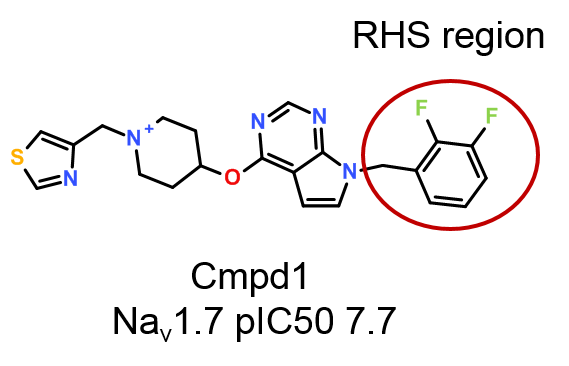
In this case study, the activity cliff summary method in Activity Atlas was used to analyze the SAR of the right-hand side region (RHS, Figure 1) of a small data set of published3 pyrrolopyrimidine antagonists of voltage-gated sodium ion channel Nav1.7, a major regulator of human pain and an attractive target for the development of new and effective pain therapeutics.
The objective is to prove the usefulness of Activity Atlas maps in understanding the electrostatic, hydrophobic and shape features underlying biological activity in those cases where structural information about ligand-target interaction is not easily accessible.
The detailed SAR analysis of the other regions of the pyrrolopyrimidine antagonists will be presented in a future case study.
A small data set of 62 pyrrolopyrimidine Nav1.7 antagonists (Figure 1) originally published by Chakka et. al.3 was downloaded from ChEMBL.4
Nav1.7 pIC50 values for this data set span a 3-fold range from 4.4 to 7.7, with an even distribution shown in Figure 2. The data set includes five very weakly active
The alignment workflow shown in Figure 3 was applied to align the 62 compounds in the data set.

The use of a 3D similarity metric in Activity Atlas requires (as with 3D-QSAR) the generation of alignments for all compounds and is sensitive to misalignment and
alignment noise. The choice of a sensible conformation for the reference structure may be critical in those cases where no experimental information is available about the bioactive conformation of the ligands.
The low energy conformations of Cmpd1 were accordingly visually inspected, to select a small number of low energy conformers representative of its conformational space. These conformers were used as the reference structure in separate Forge projects to develop alternative alignments for the training set and generate distinct Activity Atlas models. These models were then checked for consistency, and further validated by exploring the detailed SAR of the data set with Activity Miner5, a module within Forge and Torch6 providing rapid navigation of complex SAR.
The training set compounds were aligned to each low energy conformation of Cmpd1 by Maximum Common Substructure using an ‘accurate but slow’ set-up for the conformation hunt:
Activity Atlas models are calculated following a probabilistic approach which takes into account the probability that a molecule is correctly aligned.
This is done by associating a weight with each alignment based on its similarity score. Alignments with similarity higher than a certain threshold (which can either be automatically calculated by Forge, or manually defined by the user) are fully trusted. Alignments with similarity lower than the low similarity threshold are not trusted and discarded. Linear scaling is applied to calculate a weight to alignments which have an intermediate similarity score.
Each run of Activity Atlas performs three types of analysis: average of actives, activity cliff summary and regions explored analysis.
In this case study, the activity cliff summary analysis was used to explore the SAR of the 62 pyrrolopyrimidine Nav1.7 antagonists. This analysis helps you pinpoint the critical regions of SAR, providing a visual 3D summary of the activity cliffs for the data set derived from the Activity Miner module. The method is described in detail in the ‘Generating Activity Atlas models’ section of the Forge manual.
The results of the activity cliff summary analysis for the phenyl ring on the RHS starting from representative low energy conformations of Cmpd1 are shown as 3D maps in Figure 4.
All the models give consistent results, and provide clear indications about the electrostatic, hydrophobic and shape features underlying Nav1.7 activity, as explained in detail in Figure 5.
Small halogens in the ortho and meta positions of the phenyl ring on the RHS of the molecule improve Nav1.7 activity, as shown by the negative electrostatic field (in cyan in Figure 5) and the associated areas of favorable shape (green areas). Areas of favorable/unfavorable hydrophobic interaction are not shown for clarity as they overlap largely with those of favorable and unfavorable shape.
Substituents which generate a more positive (or less negative) electrostatic field (in red in Figure 5) in the para and second meta positions are beneficial for activity.
Steric bulk in the para position (magenta areas) instead is detrimental for Nav1.7 activity.
Finally, electron-withdrawing substituents which generate a more positive (or less negative, in red in Figure 5) electrostatic field below the plane of the ring are also beneficial for Nav1.7 activity.
These general trends were investigated in more detail by means of activity view maps calculated and displayed using Activity Miner.
The activity view shows a focus compound surrounded by its nearest neighbors according to the chosen similarity metric (Figure 6). In this view the height of each wedge corresponds to the ‘distance’ between the pair: a smaller wedge reflects very similar compounds.
The color of the wedge reflects the direction the activity is going: red means the activity is decreasing; green means the activity is increasing between the pair. The shading echoes the disparity, which relates to how steep the activity cliff is. The result is a focused view of the SAR around a particular compound.
Figure 6 shows the activity view around o-F-phenyl (pIC50 7.7), one of the most potent compounds in the data set.
Starting from the o-Cl substituent and going clockwise, it can be seen that replacing the o-F substituent in the focus compound (pIC50 7.7) with o-Cl (pIC50 7.3) or o-Me (pIC50 7.4) has a very slight detrimental effect on activity, as these substituents are associated with a less negative electrostatic field.
Introducing a second F in the meta position does not impact activity (pIC50 7.7), while the unsubstituted phenyl ring is much less active (pIC50 6.6), as it lacks the favorite small halogens in ortho, meta.
Replacing m-F with m-Cl again does not impact activity.
The introduction of a second o-F substituent (pIC50 6.8) instead causes a drop in activity, an effect not highlighted by the activity cliff summary, as only one example of ortho, ortho disubstitution is available in the data set.
Replacing o-F with o-CF3 (pIC50 7.1) causes a modest drop in activity, which is difficult to explain in terms of electronic effects: this compound is possibly an outlier to the general trend shown by the activity cliff summary maps.
Removal of the o-F substituent causes a drop in activity, as can be seen for m-F (pIC50 6.9), m-Me (pIC50 7.1) and m-Cl (pIC50 7.1).
Finally, the lack of small halogens in the ortho and meta positions, together with the introduction of unfavorable steric bulk in the para position, causes the dramatic drop in activity in p-F (pIC50 5.1): this substituent is also associated with a more negative electrostatic field.
In this case study, Activity Atlas and Activity Miner were successfully applied to decipher the SAR of the right-hand side phenyl ring of a series of voltage-gated sodium ion channel Nav1.7 antagonists, starting from a very limited amount of SAR data and no available crystallographic information about the bioactive conformation.
The activity cliffs summary in Activity Atlas was used to get an overview of the SAR landscape, focusing on the prevalent SAR signals.
Activity Miner was used to drill down into the Activity Atlas maps to understand subtle molecule-to-molecule structure-activity changes and identify potential outliers.
The two methods used in combination were able to quickly identify and decipher the most relevant features underlying protein-ligand interaction.
The information derived from this analysis can be of invaluable help for drug discovery projects to inform design decisions and help prioritize molecules for synthesis.