In December 2016 I attended the SCI Protein-Protein Interaction symposium. Armed with Cresset’s powerful ligand centric molecular modeling suite Forge, and an embryonic version of our new structure-based design application, Flare, I was keen to see what could usefully be done with PPI’s.
Prof. Richard Baylis (University of Leeds, UK) presented new data on the interaction of N-MYC with Aurora A. N-MYC is a disordered multi-domain protein with a host of interaction partners. Dysregulation of N-MYC has been linked to a range of cancers. N-MYC is short lived in-vivo and its usual fate is to be ubiquitinylated and degraded. Binding with Aurora A protects N-MYC from this process allowing its various tumorogenic affects to persist. The Baylis group provided the first x-ray evidence showing how N-MYC interacts at an allosteric site of Aurora A which stabilises an active conformation of the Kinase (figure 1).
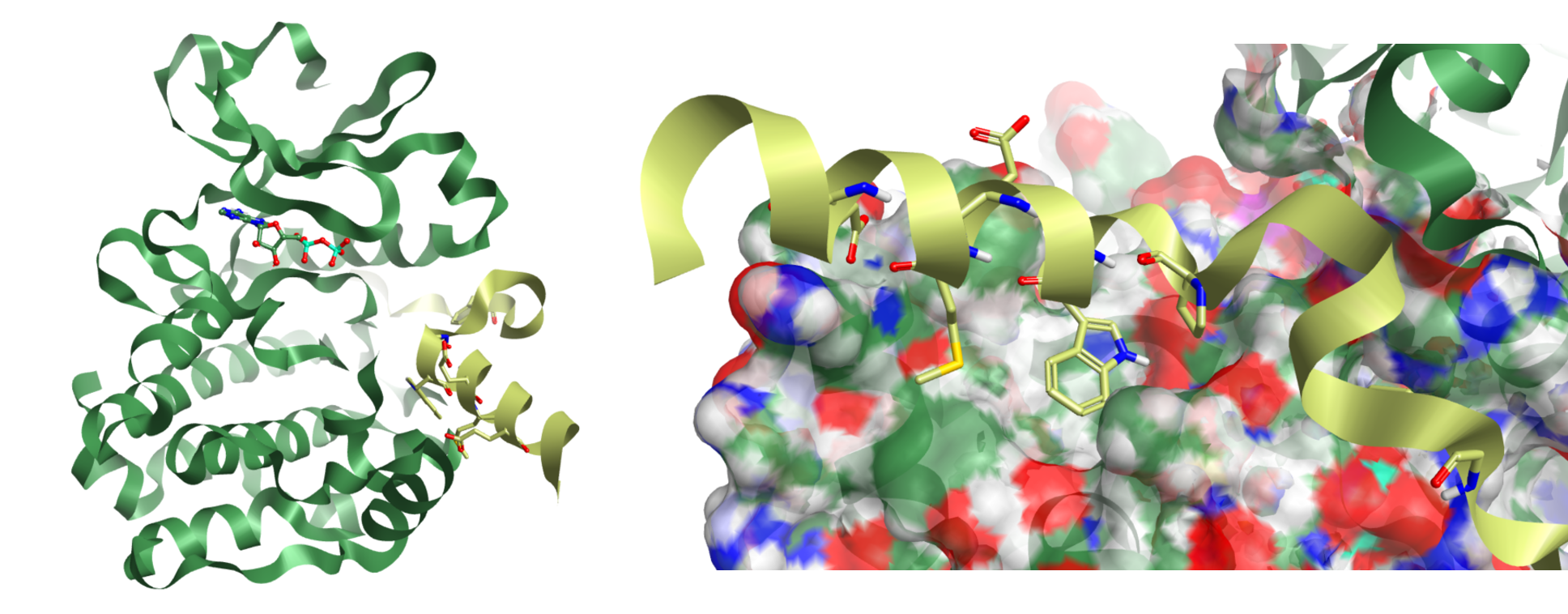
Figure 1: Aurora A kinase with N-MYC – light green (left), and detail of the N-MYC short helical domain 74-89 (right).
Baylis suggested that DFG-out inhibitors of Aurora A provide distortions of the kinase that would prevent MYC binding, conversely, inhibition with ATP competitive inhibitors would not. Evidence of potential beneficial effects of the former type of kinase inhibitor, but not the latter, may be explained by this fact and led to the suggestion that this may be an effective therapeutic strategy for some types of cancer such as neuroblastoma.
An alternative computational strategy, which occurred to Cresset at the time, was to employ a structure-based approach; to furnish molecular designs that could directly prevent this protein-protein interaction. For this purpose, an initial analysis of the surface interaction, including both electrostatic and lipophilic hot-spots, would be vital.
During the talk, I used Flare to quickly download the relevant PDB file (5G1X) and to load the protein coordinates directly into the application. An automated protein prep protocol (build-model) was used to refine the pdb structure before generating the surface interaction maps, using Cressets proprietary XED force field (figure 2).
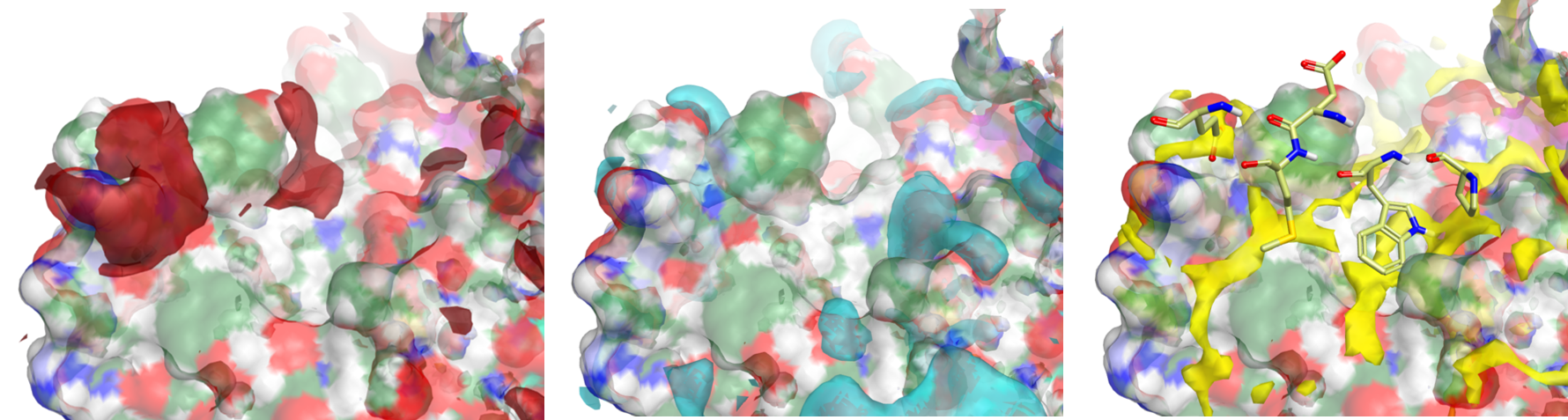
Figure 2: (A) Positive protein electrostatic isopotential surface of Aurora (left), negative protein electrostatic isopotential surface (center), and neutral isopotential surface with some key residues of N-MYC (right).
These isopotential maps show discrete positive (red), negative (blue) and neutral (yellow) surface regions that represent key interactions sites between N-MYC and Aurora A which allowed the assignment of the N-MYC residues on which to focus. The N-MYC protein was similarly used to generate and visualise the complimentary fields – as the other component half of the PPI (figure 3).
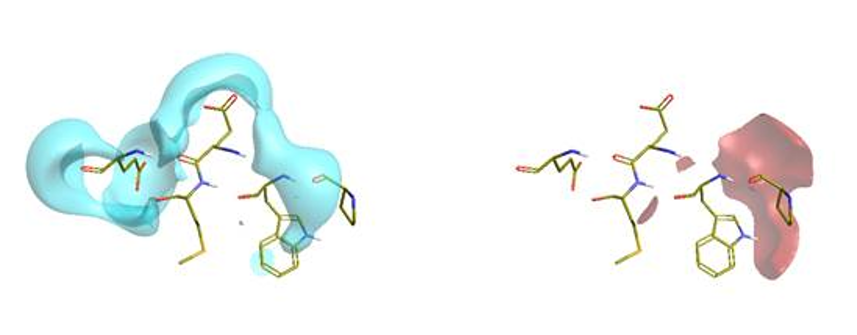
Figure 3: Negative protein electrostatic isopotential surface of N-MYC short helical region (left), and positive protein electrostatic isopotential surface of the same (right).
In keeping with other known PPI’s such as the MDM2 system, in the short helical domain (N-MYC 74-89) residues Met81 and Trp77 were identified as key lipophilic contacts. Much of the rest of the helix is largely for structural integrity and for stabilising solvent except for the NH of Trp77 and Glu84, which provide additional polar contacts, the latter capping an adjacent helix from Aurora. Further along the N-MYC peptide, towards the N-terminus, Pro74-Pro75 motif (figure 1) marks a change in sec. structure leading to another lipophilic contact Val61 and another polar contact Ser64 (not shown).
We can exploit this information to generate chemical starting points, once each important set of residues is identified and mapped. Thus, from the 3D shape and detailed electrostatic information we can conduct de-novo design experiments to furnish ideas for synthesis, or use virtual screening (Blaze) to search for commercial compounds to purchase and test.
Since the distance between the two main hot spot regions was not ideal (27 Ang. Val61 to Trp77) and although linking them might have been possible using a fragment linking or growing technique e.g., using Spark (Using Cresset’s Spark to grow and link distant fragment hits with sensible chemistry), we chose to tackle them independently with a de-novo design technique. I used the key residues Pro75, Trp77, Glu80, Met81 and Glu84 from the short helical domain as a molecular reference. We used this reference to score our molecular ideas against, and to optimize them via iterative ‘molecular design > alignment > scoring’ cycles in Forge. This powerful technique scores 3D shape, electrostatics and protein steric clashes whilst simultaneously calculating and/or filtering in-silico physiochemical properties. This method as described is limited only by the imagination of the user. In conjunction with Spark as the idea generator however, the limit is set only by the availability of appropriate fragments in the Spark databases – which is a substantial resource.
Later, when we returned, we also ran a virtual screening test on this system using Blaze. Results of this quick virtual screen against a sub-set of the ChEMBL database are shown below (figure 4).
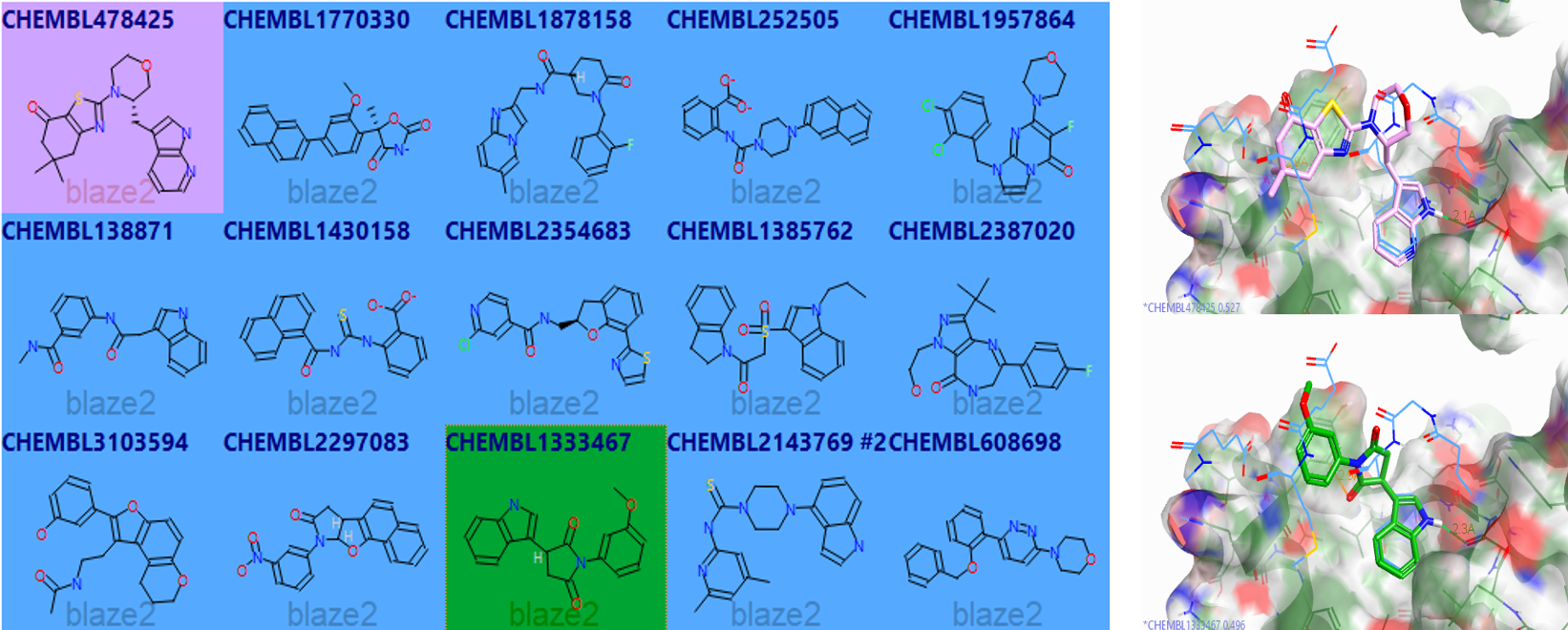
Figure 4: Forge ‘tile view‘ of example diverse 2D output results of the virtual screen using Blaze against a sub-set of ChEMBL (left) and 3D alignments of two of these (pink and green sticks) against the reference N-MYC peptide (blue lines) bound to Aurora A (Forge screenshot).
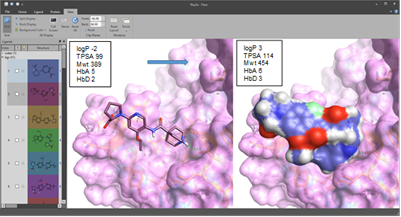
Although some of the Blaze examples retrieved were interesting, very good considering that this was a very small set <200k compound DB, it appears that good shape score and field score were not generally observed simultaneously. The ‘new’ addition of ‘pharmacophoric atom features in Blaze’ ensured we retrieved some of the key contacts such as the indole H-bond. However, we felt that design was probably the best way to address achieving the precise set of contacts we were looking to mimic. Afterwards, I expanded on the ‘initial’ de-novo design ideas and provided around 20 further designs which had more reasonable properties and synthetic tractability (figure 5).
A powerful combination of cutting edge ligand and structure-based modeling
Figure 5: Flare screenshot of the structure of an initial idea (left) superimposed on N-MYC hot spot residues, plus its calculated properties, and (right) a space filling model of a further example with superior properties, improved fit, better synthetic tractability and … an IP position.Although this is only a thought experiment (until the point at which any of these molecular designs are synthesized and tested) this illustrates how the powerful combination of both ligand centric and structure-based techniques in Flare, Forge, and perhaps also Spark, could be used to generate specific ideas that address the types of challenges presented by PPI’s or fragment enabled drug discovery projects. This is not untypical, in terms of a portfolio of tasks we might suggest to Cresset Discovery Services clients.
Download an evaluation of Flare, Forge, Spark and Blaze, or contact us to find out how Cresset Discovery Services can enhance your project with insightful and creative delivery of powerful molecular modeling.




