It’s a long journey from hit to lead, and the path is called optimization.
Your discovery program started with some promising leads from a high throughput screen. The biology has been done and the chemistry has shown that there is an effect on the biochemical assays. You have a series of hit molecules, a set of required parameters, thousands of data points, a team of chemists, a budget and a deadline. Now the optimization work begins.
In silico methods streamline the optimization process by giving you more understanding of your target and your hits, and by making it easier to manage your data. There are methods to:
- Identify any gaps in your data so that you can decide what further work needs to be done. The more you know, the greater choice of intelligent steps you have.
- Make sure you have the best possible understanding of your molecule-target interaction, giving you as many optimization options as possible.
- Balance diverse properties. Visualization and multi-parameter optimization tools can transform your ability to understand the impact of changes across different compounds.
- Stay in the active window as you make minor changes. Building an activity pharmacophore helps you to understand how far you can take the changes.
- Escape any liability from toxicity or pre-existing patents. Fragment replacement methods can be invaluable for moving to new areas of chemical space and adding new ideas and directions to your research.
Are there gaps in your data?
The optimization decisions you can make depend on the data you already have.
Activity Atlas, a component of Forge, summarizes the SAR for a series into a 3D model that can help you find any gaps in your data. You can calculate:
- Activity Cliff Summary: What do the activity cliffs tell us about the SAR?
- Average of actives: What do active molecules have in common?
- Regions explored: Where have I been? For a new molecule, would making it increase our understanding? This analysis also calculates a novelty score for each molecule.
This approach is also helpful in looking at toxicity and other liabilities. For example, you may be optimizing a molecule that was identified from a screen. It is active, but has some undesirable chemistry. If you can understand as much as possible about the SAR, electrostatics and shape you are more likely to discover a better way to escape a liability. The more you know, the greater choice of intelligent steps you have.
Figure 1: Activity Atlas condenses your structure-activity data into highly visual 3D maps that inform the design and optimization of new compounds.
Understand the ligand-protein interaction
Understanding how your hits interact with the target helps you to optimize the affinity of your compound. In structure-enabled projects Flare, our new structure-based design application, can be used to analyze the protein-ligand system, calculate the energetics of ligand binding and analyze the water stability and energetics.
Without knowledge of the target structure, you may need to go back and deduce ligand-protein interactions from your hit so that you have a clearer understanding of the binding mechanism. This knowledge makes it far easier to optimize the affinity of your compound.
You get an extra level of insight to this process with Cresset techniques. Our electrostatic, hydrophobic and shape based analyses make it clear which chemical changes can have the largest biological impact.
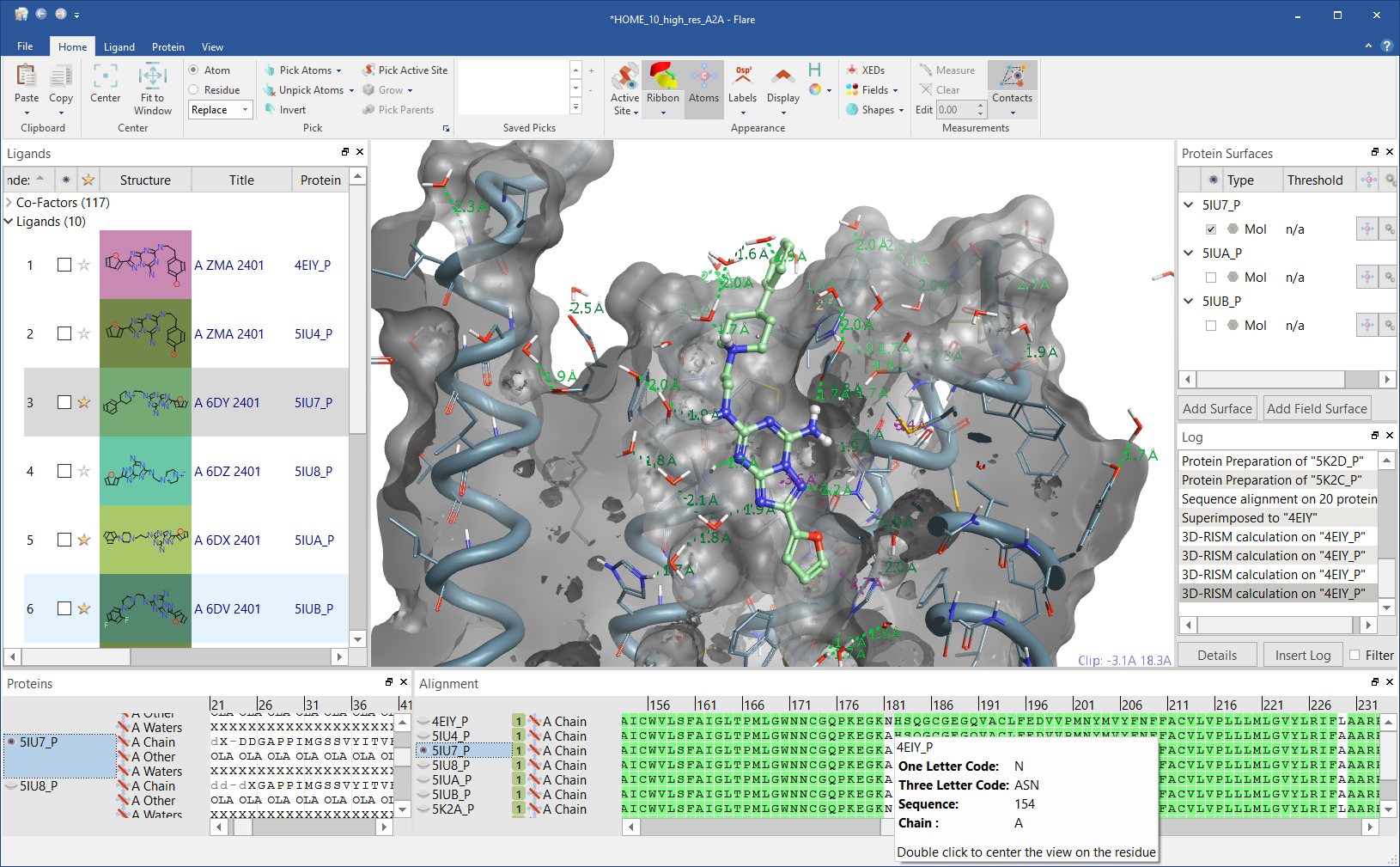
Figure 2: Flare GUI.
Visualization tools help with multi-parameter optimization
Compounds that come out of the screening process generally have weaker potency than is required, so one of the first tasks is to increase the activity. This is usually done by making small changes to the molecule and testing their effect. However, there is a range of properties besides activity that need to be optimized.
Firstly, drug molecules need to be stable and small. Larger molecules are more likely to have off-target effects and have more problems travelling through membranes. They are also likely to have more complicated chemistry, making them harder and more expensive to synthesize. One of the key steps in optimization is to retain as many components as are required to make the compound active, but no more. The difficulty is that when you change one property you tend to change others.
Computational visualization for multi-parameter optimization shows you how the changes you are making affect other molecular properties. For example, you may want to simultaneously optimize the polar surface area and the LogP. The Torch and Forge radial plots can be set up to define acceptable project ranges for project data and in silico–calculable properties.
Contact Cresset Discovery Services for a confidential discussion about how we can streamline your optimization.

