Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
The latest version of BlazeTM, our virtual screening platform is now available. V10.3 introduces pharmacophore constraints to enable you to find the best possible new hits and leads. Alongside pharmacophore constraints, we’ve added additional similarity metrics and updated the user interface.
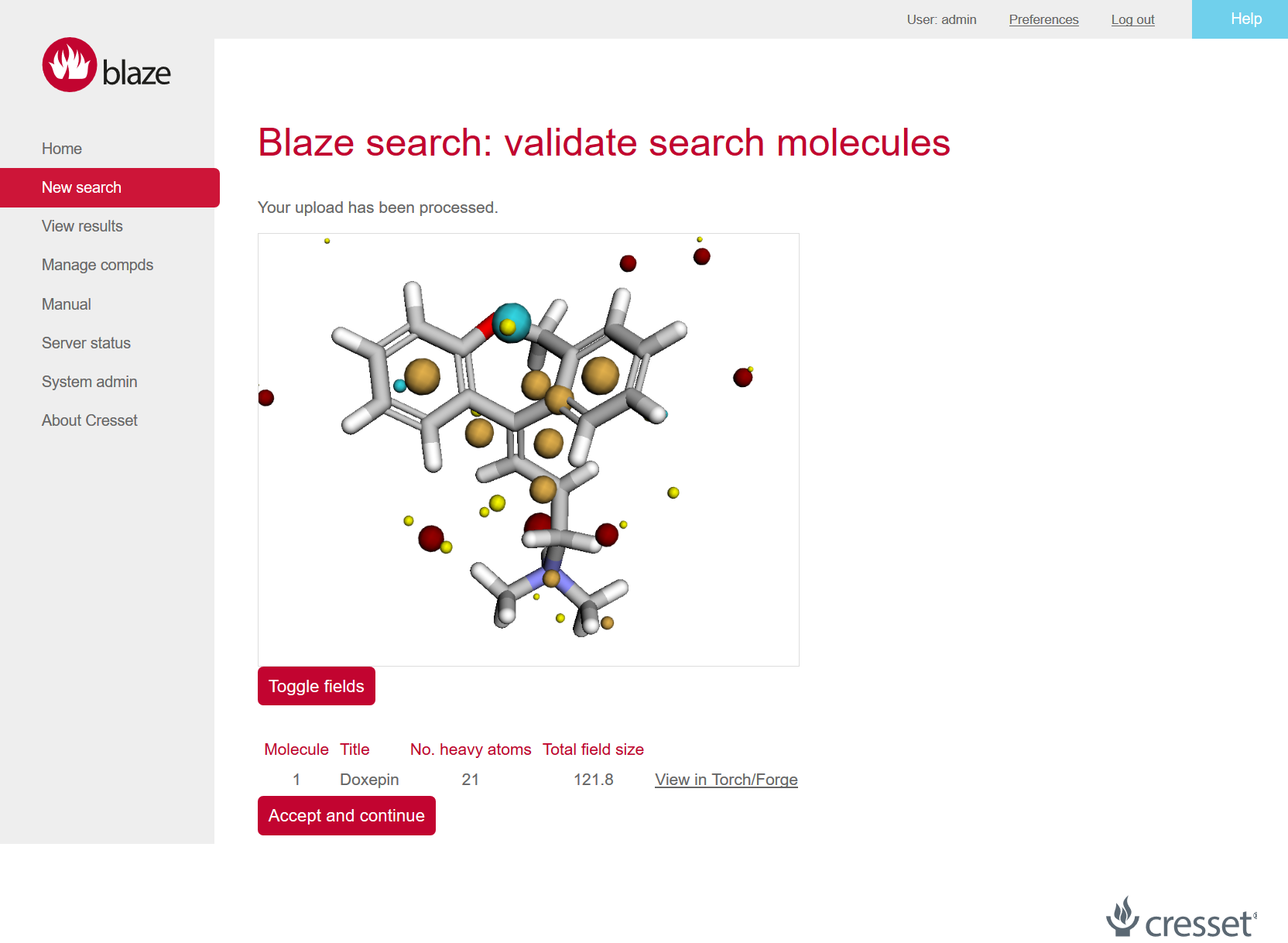
Figure 1: Blaze has a new look that includes WebGL views of the search molecules
In previous versions of Blaze we have enabled the setting of Field constraints. These work by down-weighting any result that did not have a specific electrostatic or hydrophobic field at a location you specify. V10.3 enhances this capability by giving you the ability to add a specific atom as a pharmacophoric feature that must be matched by an atom of a similar type in the results. The effect of this is to provide you with a mechanism for ensuring that the results that you get from your virtual screening experiment fit with the known SAR or with your expectations. For example, using pharmacophore constraints you can ensure that all results retrieved from a virtual screen for new kinase hinge binders have the donor-acceptor-donor pharmacophore motif. This differs from field constraints by the severity of the match required. Using field constraints a donor can match other motifs that also express positive electrostatics – such as electron deficient aromatic C-Hs where a pharmacophore feature would only match hydrogen atoms attached to heteroatoms.
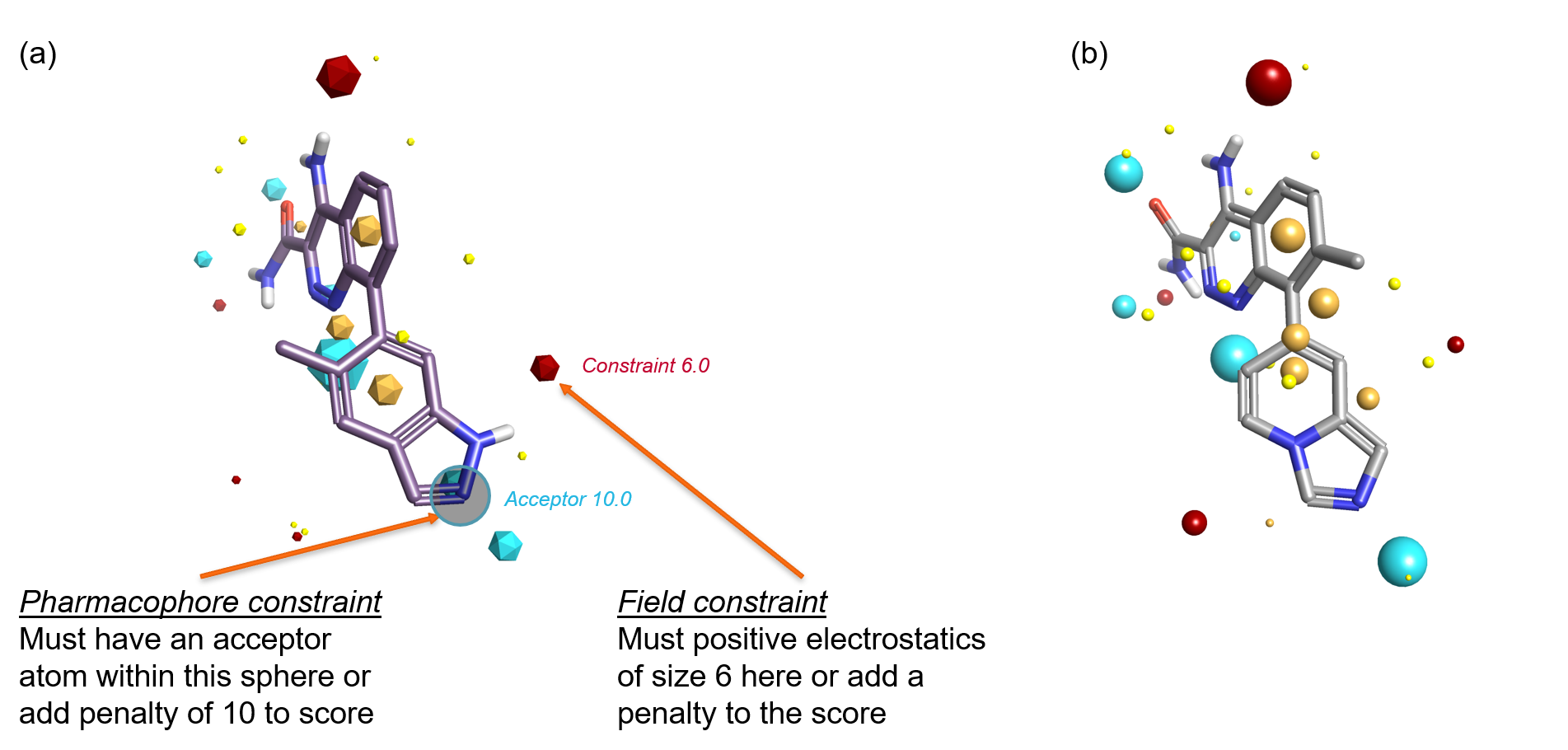
Figure 2: (a) Ligand from PDB 4Z3V with field and pharmacophore constraints added. (b) Active BTK inhibitor that satisfies both constraints. Note that the aromatic hydrogens match the field constraint but would not have matched a pharmacophore constraint placed on the indazole NH.
We tested the new pharmacophore constraints using a selection of kinase targets taken from the DUD dataset. We applied constraints to the hinge binding motif of each query molecule and studied the retrieval rates. Overall we found an average improvement of around 0.13 in ROC-AUC across the tested targets which represents a reasonable gain given the deficiencies in the dataset.
The ability to constrain result molecules to those that fit a specific pharmacophoric feature is very powerful. However, we advise caution – there are many known actives that do not necessarily contain a specific pharmacophore. This is highlighted in the BTK example above but can also be seen in kinases. For example, the CDK2 ligand from PDB 2uzl, although less active than some chemotypes, lacks any of the classic pharmacophore features associated with hinge binding and hence would not be retrieved by a query with constraints on these features.
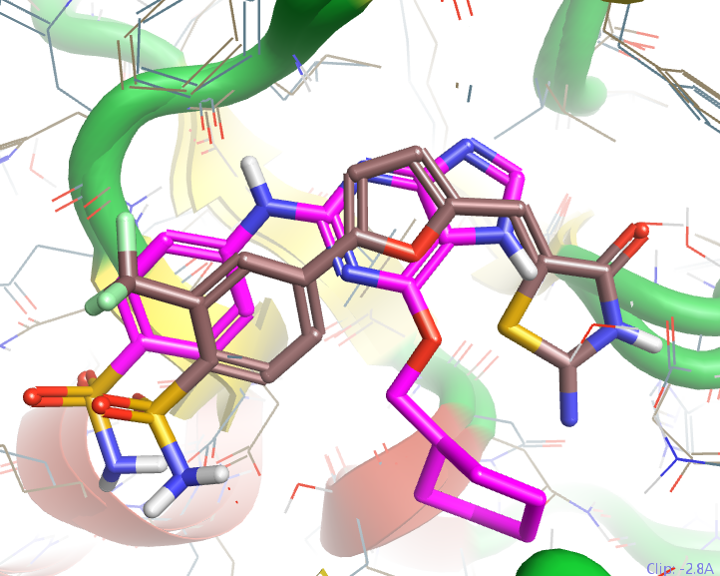
Figure 3: Overlay of equivalent C-alpha atoms of PDB 2uzl (ligand in brown) and PDB 3c6o (ligand in pink). The 2uzl ligand lacks all of the classic pharmacophoric hinge binding motifs.
Perhaps the most interesting aspect of the new pharmacophore constraints is in the application to virtual screening for covalent inhibitors. These enable you to specify that the retrieved molecules must contain a electrophillic center at same position as in your query. This works in exactly the same way as the traditional H-bond donor, H-bond acceptor type of pharmacophore constraint. In the ligand alignment algorithm we downweight any alignment where an electrophile is not overlaid with the constrained atom. This could be especially useful when screening large virtual libraries or other custom collections where the standard filters for screening collections are not appropriate. As well as electrophiles, we have an definition for metal binding warheads which, again, should help find a richer set of compounds for wet-screening than was previously possible.
The Blaze interface has a new crisp look that emphasizes the easy-to-use nature of the web interface. Unlike other virtual screening algorithms, Blaze is a complete system that enables easy compound and collection management combined with user and project based permissions. All of this is accessed through a web interface that has a wizard approach to experimental setup.
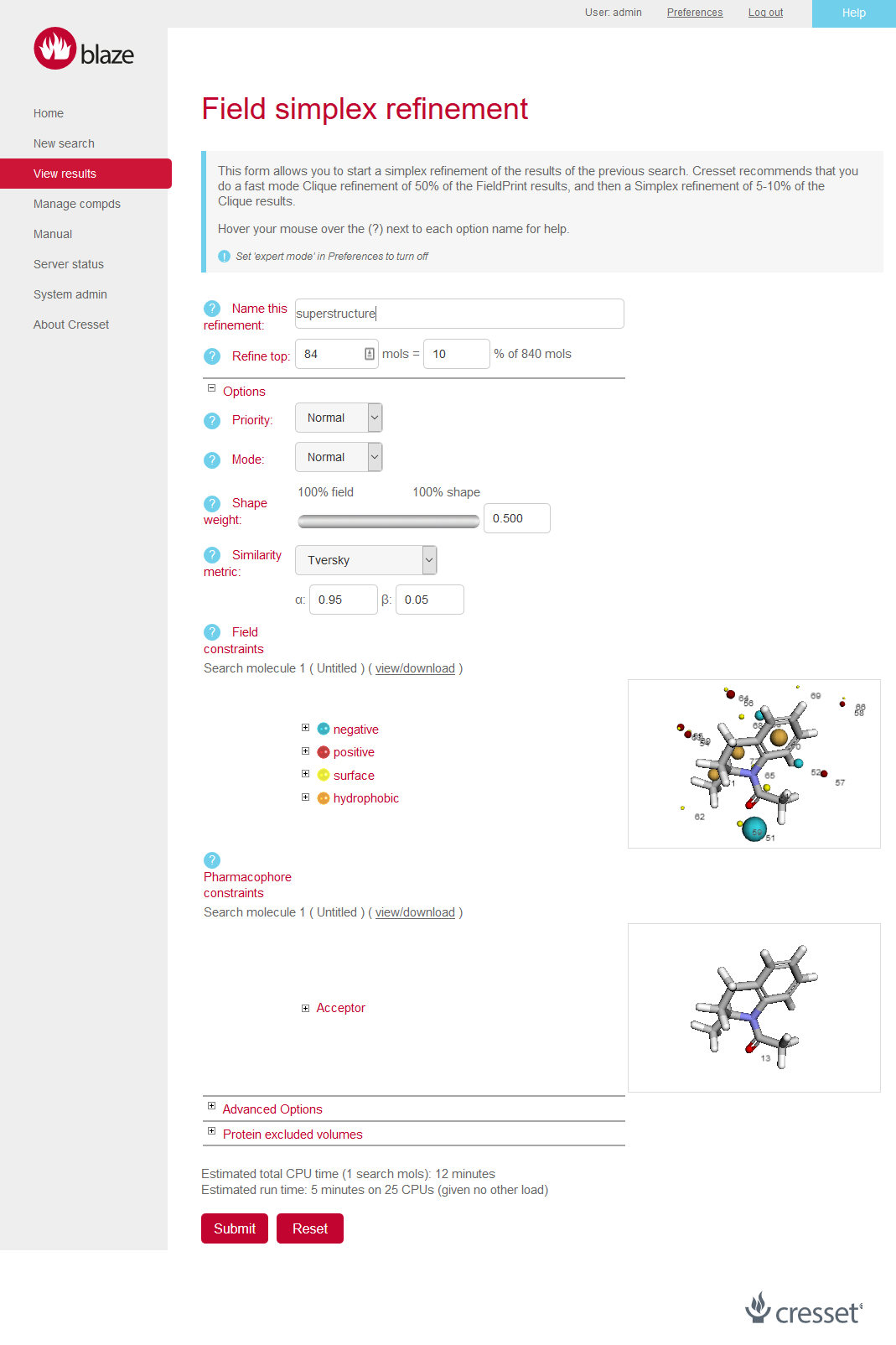
Figure 4: The New Blaze interface is cleaner with color coordinated help and prompts.
The web interface is not the only way to use Blaze. Our desktop applications Forge and Torch use Blaze’s REST API to submit searches and retrieve results giving you access to the power of Blaze from your desktop. However, the REST API can be incorporated into virtually any other application and we provide Pipeline pilot protocols, and example KNIME workflows to show how to search and manage compounds from these workflow solutions.
All the new science released in Blaze V10.3, described above, is available through the REST API.
Blaze is available as software for installation on your internal cluster, as an Amazon Machine Image that will run within your Amazon deployment or for rental on a per project basis using our Blaze Cloud installation. Contact us for more information.