Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
V10.5 of ForgeTM, the powerful computational chemistry suite for understanding structure activity relationship (SAR) and design, is now available. This release introduces significant enhancements to molecule alignment, plus the new Conformation Explorer, to visualize and inspect conformational populations. Also included are a large number of GUI styling and usability improvements.
Molecule alignment is the core experiment in Forge. It is key to developing robust qualitative or quantitative SAR models, building FieldTemplater pharmacophore hypotheses, understanding the design of new compounds and small scale virtual screening experiments (for larger scale virtual screening use Blaze). V10.5 enables fine-tuning of alignment results by introducing appropriate constraints, an optimized substructure alignment algorithm, and new similarity scoring options.
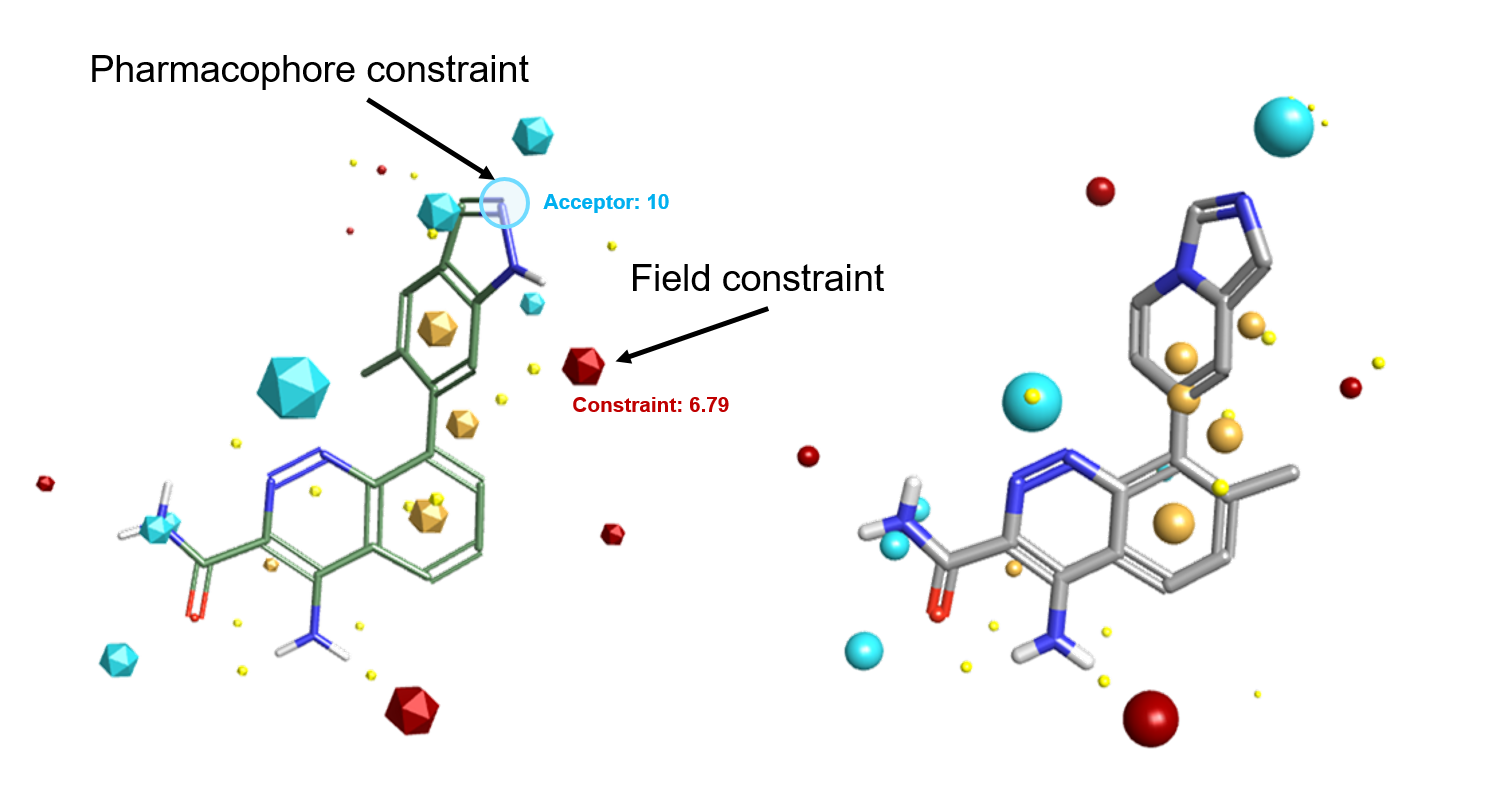
Field and pharmacophore constraints bias the alignment algorithm by introducing a penalty which down-scores results that do not satisfy the constraint. This provides you with a mechanism for ensuring that the results that you get from your alignment experiment fit with the known SAR or with your expectations.
With field constraints, you can specify that a particular type of field must be present in the aligned molecule. For example, you may want to a constrain a positive field where you want an interaction but this can be matched by both H-bond donors and other electropositive features such as the aromatic hydrogens in the example below.
V10.5 introduces the new pharmacophore constraints, which ensures that your desired pharmacophore features (e.g., Donor H, Acceptor, Cation, Anion) are matched by an atom of a similar type in the alignment results. A pharmacophore constraint can be used when you are certain that a particular interaction requires transfer of electrons (as in H-bonding or metal binding) in addition to the electrostatic character of the interaction.
Pharmacophore constraints introduce a tighter constraint on the alignment than a field constraint. Where field constraints allow matches across chemical features, pharmacophore constraints are limited to matching specific functional groups (e.g., specific donor-acceptor interactions): alignments that do not place a suitable atom on top or close to the constrained atom cause a penalty to be applied to the score. However, pharmacophore constraints in Forge V10.5 go beyond traditional H-bond donor/acceptor definitions to include, for example, covalent centres and metal binding motifs giving the ability to ensure that key warheads always align in the correct positions.
While field and pharmacophoric constraints are a powerful way of fine tuning alignment results, we recommend that they are using sparingly, as they will be introducing a bias in your Forge experiment. E.g., introducing a pharmacophore constraint on the indazole NH of the PDB 4Z3V ligand in Figure 1 – left would not have matched the aromatic hydrogens of the active ligand in Figure 1 – right.
Enhancements to alignment and scoring, accessed from the advance options panel, include:
Molecular conformations are central to Forge. The conformation hunter does a good job of generating a diverse range of energetically accessible conformations. V10.5 gives you the opportunity to more easily inspect the conformations generated for your molecules, enabling you to interact with and edit the populations.
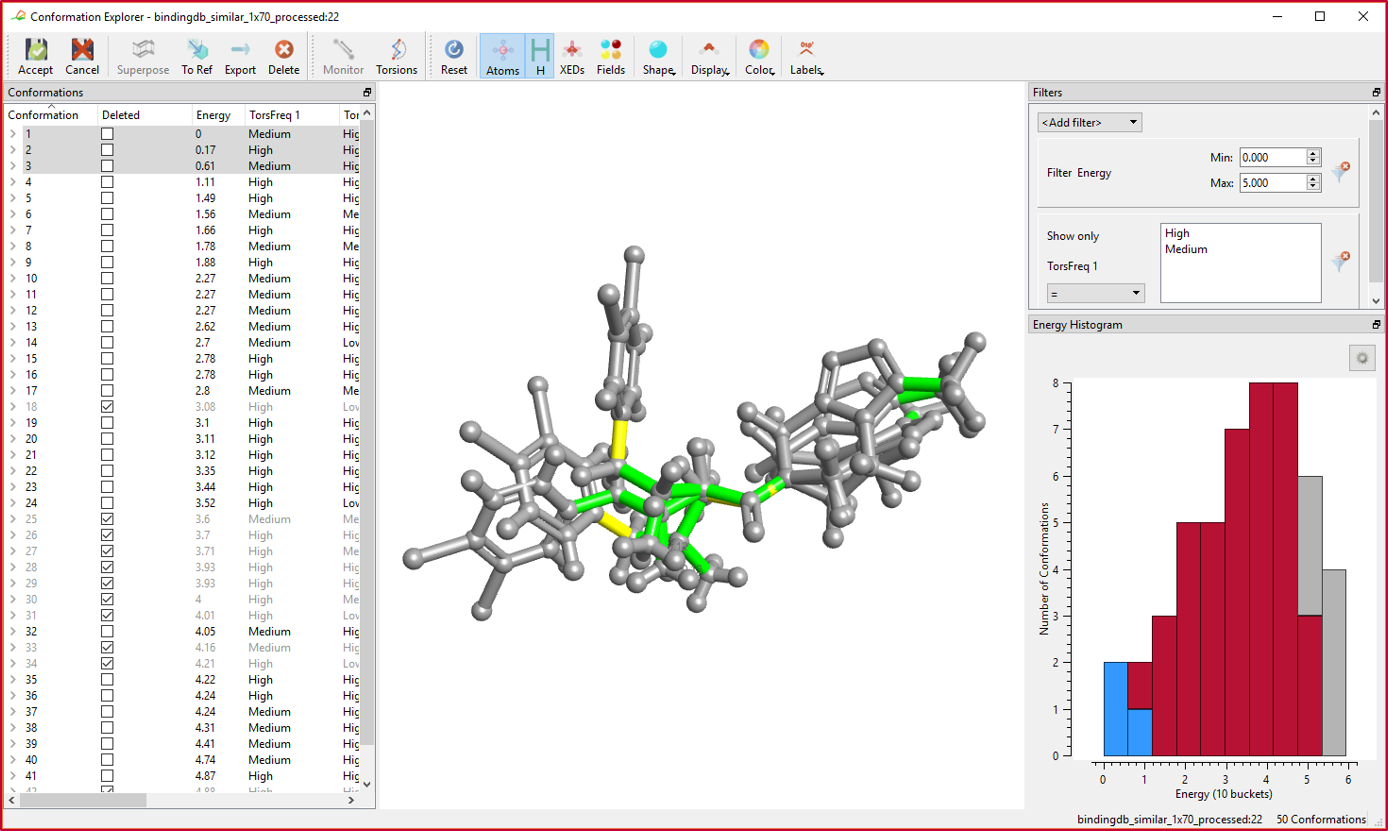
In the new Conformation Explorer, you can inspect a set of conformations with respect to energies, measured distances/angles/torsions, as well as calculate the CSD torsion frequency for each rotatable bond to assess the feasibility of the generated conformations.
Conformations are listed in order of increasing relative conformational energy. Unrealistic conformations or those which are not deemed interesting can be selected and removed from the conformation population for that molecule. Preferred conformations can be promoted to the reference role in Forge with the click of a button.
CSD torsion frequencies can be calculated for all rotatable bonds. These are based on the Torsion Library which contains hundreds of rules for small molecule conformations derived from the Cambridge Structural Database (CSD) and curated by molecular design experts. CSD torsion frequencies are useful to highlight cases where the torsion angle in a calculated conformation is not one that is frequently observed in the CSD, and accordingly is a possible cause for concern.
Distances, angles and torsions can be measured for each conformation and those values can be used for filtering or generating a histogram plot.
Conformation energies can also be plotted in an interactive histogram plot. In Figure 2, the column or bucket with the blue highlight reflects the current conformations shown in the 3D view; the grey columns or buckets reflect to conformations which do not pass the set of filters.
Conformations can be filtered by energy, CSD torsion frequency and calculated distances, angles, torsions. Smart coloring includes coloring by energy and by CSD torsion frequency.
This V10.5 release also includes a variety of additional new functionalities and improvements to the Forge interface, including:
Upgrade at your earliest convenience to try the new Conformation Explorer and pharmacophore constraints in Forge, together with the many new and improved features in this release.
If you are not currently a Forge customer, download a free evaluation.