Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
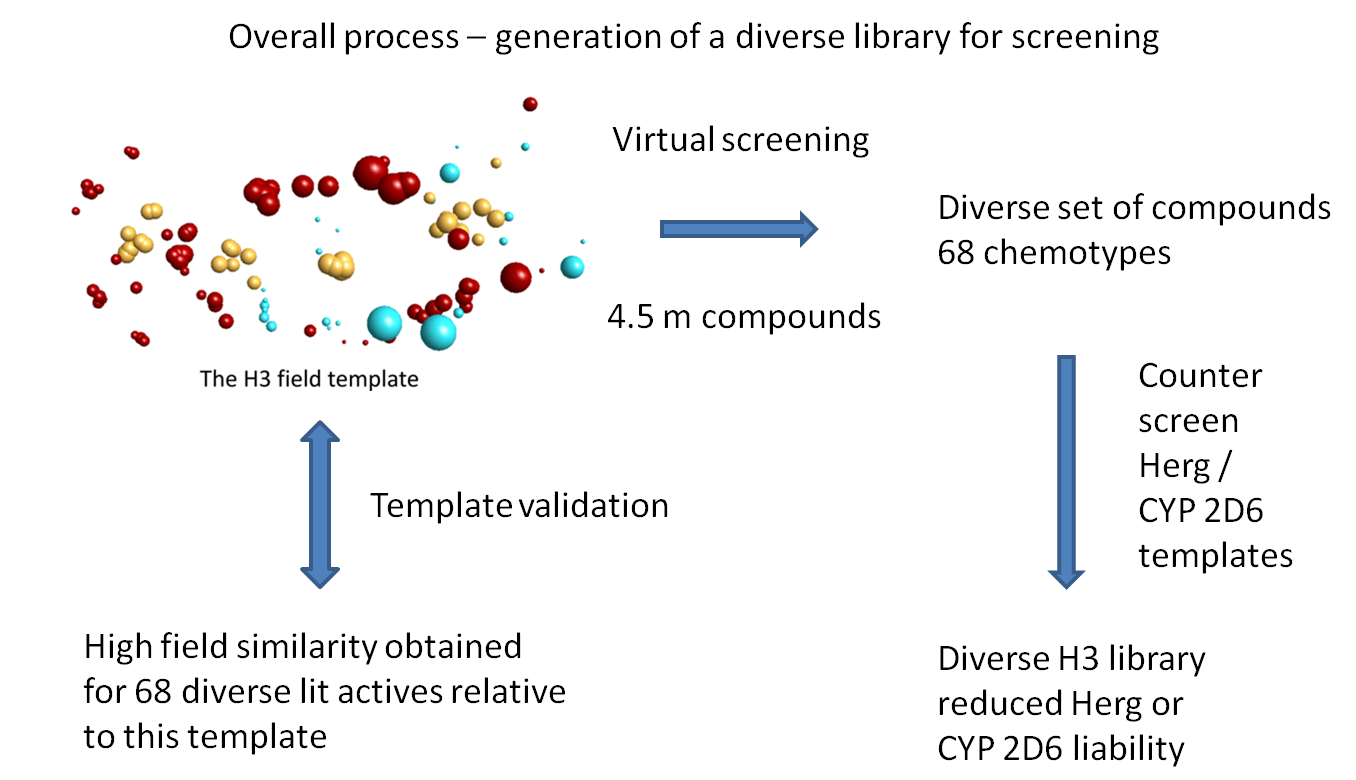
There is a delicate balance to be struck in library design between identifying all the chemical scaffolds that are potentially active, whilst retaining a manageable library of compounds that is tractable and cost-effective for routine screening. Field templates can be used in library design to predict activity of compounds both at therapeutic targets as well as at known toxicity targets such as CYP 2D6 and hERG. A range of templates can be derived and used as ‘lenses’ to counterscreen an aggregated library of compounds derived from multiple congeneric series and other sources.
FieldTemplater was used in the example shown to select to select a diverse library of potential H3 antagonists. A series of seven highly active H3 antagonists were identified from the literature and aligned in their bioactive conformations to generate a consensus field template (shown bottom). As confirmation of the predictive capability of this template, the field match score was compared against the known activity (Ki) scores of 68 further H3 antagonists described in the scientific literature and outside the original training set. A good match of fields to activity was confirmed.
The H3 template was then used to counterscreen Cresset’s 4.5M compound collection to identify potential H3 antagonists. A large number of matches were identified, with 68 distinct chemical scaffolds. Since chemical scaffolds that can be expected to show liabilities for serious off-target or toxicity effects should be avoided, the compound matches were also screened against toxicity Field templates for CYP 2D6 and hERG activity. Approximately 4% of the compounds were rejected due to potential 2D6 toxicity and a further 8% due to potential hERG toxicity.
Field based methods can also be used to predict novel bioisosteric compounds that will exhibit the same activity when key fragments of their structure are replaced. Such a tool was used to replace the central core as an alternative library method in order to generate a novel scaffold replacement library. The results of this analysis can be seen in the graph shown below.
The highlighted structures on the graph represent some of the most active known H3 antagonists from the literature and the blue structures represent novel compounds generated by the software. The graph shows a number of novel compounds with diverse central cores that have significantly higher predicted activities at H3 (as shown by higher field similarity score). Five of the more interesting compounds (all of which are novel and have high similarity scores) have been highlighted in yellow.
These highlighted compounds would be ideal candidates for inclusion in the final library as they combine innovation with chemical tractability and high predicted activity. Interestingly, the 2D similarity score of most of the dataset, including all of the highlighted molecules, is less than 0.7, which is a de facto cut-off for 2D based scoring methods. This means that most of these structures would be very unlikely to be considered in a traditional library design process as there would be no reliable way to predict their activity.