Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
In this series of blogs, Dr Robert Scoffin, CEO of Cresset, explores how computational chemistry is being applied to the field of reprofiling.
In my last post I talked about the straightforward ways companies can exploit their existing drugs – by reformulating, recombining and repositioning them. I briefly discussed that repositioning targets may become obvious through literature searches for side effects or off-license prescription. In this post I’ll talk about how computational chemistry can point the way to new targets for existing drugs through the use of pharmacophores.
Pharmacophores are a computational method of expressing the activity profile of a molecule. This activity profile can be used to search for other compounds with similar profiles. When you find a match you can compare the known targets for the two compounds. When two compounds with a similar profile are known to be active against different targets then you have found a promising place to focus your research.
Of course, the key to success in this area is to have built an effective and reliable pharmacophore. Firstly, it needs to be based on good science, and at Cresset we rely on field technology for this. You can read more about field patterns and Cresset’s field points here. Secondly, computational searches inevitably require simplified search times to make searching large amounts of data possible in finite time, so we need to be sure that our search criteria retain the relevant information.
When a number of compounds are known to be active against a particular target, you can look at the consistent patterns of interaction to determine the most important active sites for the protein. This is the best method for building up a field based pharmacophore that focuses on the likely active sites.
Forge is Cresset’s computational chemistry tool for building pharmacophores and screening them against compound databases. Forge takes advantage of Cresset’s ligand comparison method to align, score and compare molecules from a biological viewpoint. As you can see from the diagram below, it can lead to impressive results.
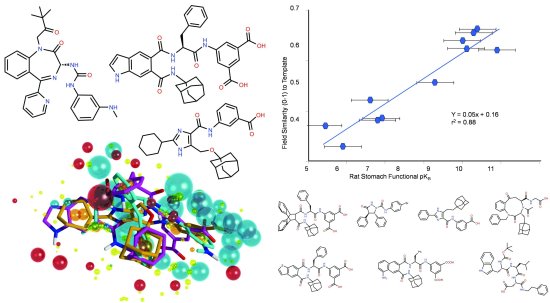
This CCK-2 pharmacophore was derived in Forge from three structurally diverse ligands. The graph shows the tight correlation between field similarity value and CCK-2 activity for seven other diverse molecules with a range of activities.