Future-proofing Cybersecurity in Drug Discovery
The pharmaceutical and biotech sectors suffer more data security breaches than any other industry, with 53% resulting from malicious activity. To protect against potential ...
News
Easter, which is nearly upon us, stems in the Latin “oestrus”, meaning “frenzy” and is the antecedent for the words “estrous” and “estrogen”. So, what better time than Easter to talk about estrogen receptor agonists and antagonists.
Estrogen Receptors
The steroidal hormone estrogen is produced by the ovaries and is the primary female sex hormone, which promotes female secondary sexual characteristics, such as breasts. During a woman’s reproductive years, the predominant form of this hormone is 17β-estradiol.
Since the early 1990s there has been much concern regarding both natural and synthetic estrogen mimics being disruptors of the endocrine system in both humans and animals. Disruption of the delicate endocrine system homeostasis can result in breast and uterine cancers, decreased fertility, precocious puberty and feminization of male animals [1].
Lipophilic estrogens easily cross the cell-membrane and once in the cytosol they bind to and activate estrogen receptors (ER) in the ovary, breast and uterus. The estradiol-ER complex controls gene expression by the protein-ligand complex binding to specific DNA sequences to activate transcription of target genes. For example, the estrogen-ERα complex can trigger the growth of new cells in the breasts, ovaries, uterus and epithelia of the efferent ducts (in males) [2].
There are several types of ER proteins, denoted α, β, and γ. The distribution of these receptor sub-types varies in different tissues – for example, ERα is in highest concentration in breast and uterus tissue, while ERα is found in bone, kidney, prostate, etc.
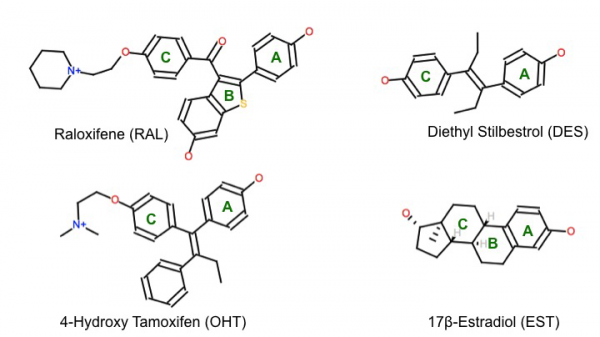
With any triggered growth of cells, the potential for transcription errors is higher, and can result in tumor cells growing and dividing at a rampant pace – this is no different for the estradiol-ERα complex’s activity. As one can imagine, a compound that acts as an antagonist to the ERα receptor should prove to be a way to treat some forms of breast cancer. Indeed this is the case with Tamoxifen and Raloxifene. Tamoxifen is special in that it is an antagonist to the ERs in breast tissue, but an agonist to those ERs in the uterus and bone tissue – hence the endometrial and bone density monitoring for breast cancer patients receiving Tamoxifen as part of their treatment protocol [3].
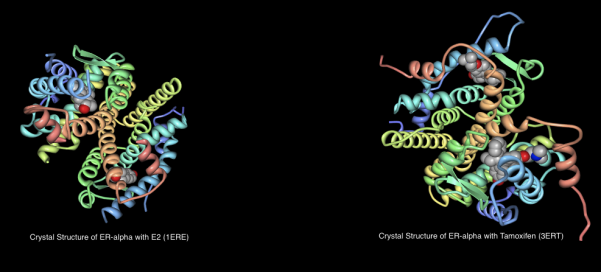
Given the availability of crystal structure information for the ERα complexed with EST, DES, RAL and OHT [5], I thought it would be interesting to run some Cresset experiments to answer the following questions:
Experiment: Forge to Determine Binding Mode
Using Torch, the following crystal structures were imported from the PDB [5]:
1ERE – ERα complexed with natural ligand 17β-estradiol (EST);
1ERR – ERα complexed with antagonist Raloxifene (RAL);
3ERD – ERα complexed with agonist diethyl stilbestrol (DES);
3ERT – ERα complexed with antagonist 4-hydroxy tamoxifen (OHT).
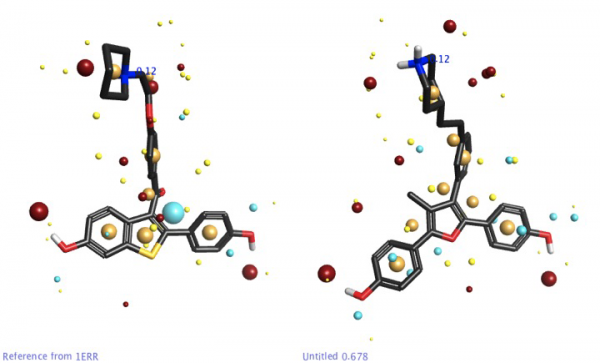
In the data preparation, only Chain A of each structure was used and all waters were deleted [6]. These ligand structures (not superposed) were transferred to Forge to calculate binding hypotheses.
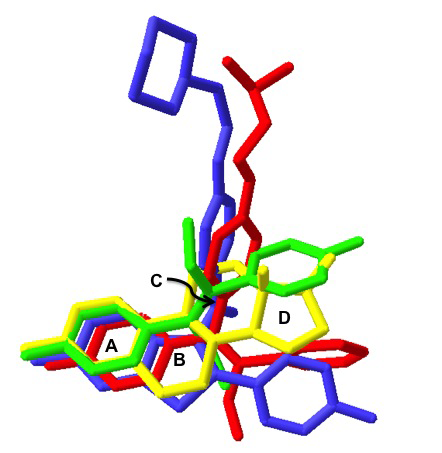
Forge is used to compare molecules based on their electrostatic, shape and hydrophobic fields in order to find common patterns. When used on molecules that have similar activities but diverse structures, this software can determine hypotheses for the conformation of the binding mode, and relative alignments of the input ligands – in the absence of protein structure information!
This experiment involves exploration of the conformational space for each of the ligands and a search for common field point patterns amongst those conformations. A common field point pattern that is generated by multiple aligned molecules of diverse chemical structure is likely to be related to the way those molecules bind to a common receptor.
The receptor information was NOT used as an excluded volume in the Forge experiment due to the conformational adaptation that occurs upon antagonist binding. Furthermore, one of the core values of Forge is that protein information is never required, which makes it a great tool for GCPR research.
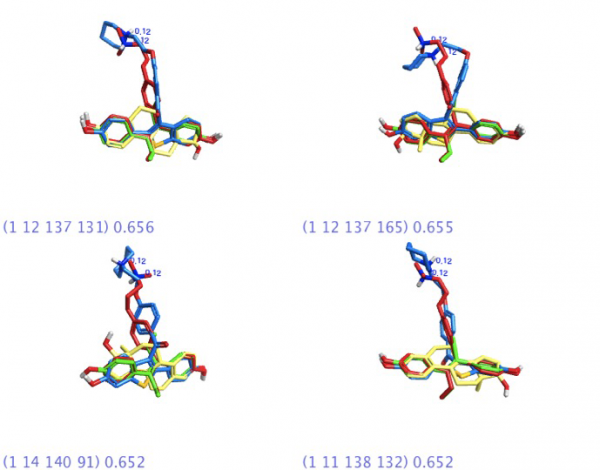
The Forge experiment on the four ligands was allowed to run with default settings. Given the high degree of structural commonality amongst these ligands, it was expected that the software would produce a large number of binding templates – there were over 300 templates that included ALL four ligands. (More structural diversity = fewer templates to examine).
Virtual Screening with Blaze on Cresset’s Cluster
On Cresset’s 120-node cluster, we have approximately 10 million commercially available compounds in commercially available libraries; for each of these compounds, up to 100 conformations are stored. Within the Blaze interface, the user has the ability to cherry pick which databases would be searched, as well as to set heavy atom ranges. Anecdotally, all 10 million compounds can be searched in about 2-3 days, depending on server load and internal priority level of the search.
In a virtual screening search for new ERα antagonist leads, it would be ideal to retain the strong binding from rings A and B to anchor the ligand tightly in the pocket. That being said, it would be really nice to get some structurally diverse leads that get us away from steroidal cores.
Based on previous success of Cresset services work focusing on virtual screening corticosteroids to find non-steroidal alternatives, I was anticipating a number of non-steroidal analogues for estrogen receptor agonists/antagonists.
blazeV10 Results – “Search”
For a virtual screen to have value, it must not only return active molecules, but also actives that have structural diversity and lead-like characteristics.
The 4-molecule template found using Forge was used as a seed for a Blaze search in the hopes of finding leads that would provide similar properties to BOTH the agonist and antagonist cores. The Blaze experiment was set to screen approximately 6 million commercially available compounds with a heavy atom range of 16-40.
In the Blaze results, there were some interesting scaffolds that were chemotypically different from the steroidal core in the template ligands, but with acceptable field point pattern matches to the four-molecule template. These cores might be considered as a starting point for lead optimization in a new chemical series.
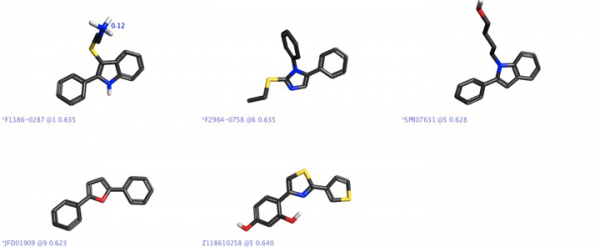
It is interesting that there is a report [8] showing that furans with basic side chains were found to be selective antagonists to ERα, with the structure shown in Figure 6 determined to be almost 70 times more selective to ERα – it was encouraging that there was a furan core in one of our “hits” shown in Figure 5.
Blaze Results – “Re-Search”
In light of the issues surrounding four-molecule templates being used for virtual screening within Blaze, I’ve since rejigged the experiment so that each of the four molecules from the template is being run individually. The results from each run will be combined and compared to identify new leads for ERα antagonism. If you want to find out what the results are, you’ll have to keep an eye on my upcoming blog posts!
Summary
In this month’s article, we discussed a workflow that encompasses the elucidation of a reasonable binding mode (pharmacophore template) with Forge, demonstrated how that template may be used for virtual screening to find diverse chemical series, and how the results can ultimately be used for scaffold-hopping and lead optimization.
We also learned about experimental set up in Blaze and the pitfalls associated with multiple-molecule templates as the virtual screen seed. Hopefully my experience will save our Blaze users time/resources by not repeating my mistake. (See Blaze tip below).
I was once told that what we do isn’t “SEARCHING”…It’s “RE-SEARCHING”. And that bit of wisdom was never truer than for this month’s work. There is so much more work to do, and I’m planning to revisit ERα again to look at the new virtual screening results in an upcoming post.
Tip for Blaze Users
Doing a Blaze search on a four-molecule template is not recommended as the search will attempt to find structures that satisfy the field point patterns of all four molecules simultaneously and that’s an awful lot of field points.
Additionally, when running on a multi-molecule template, the virtual screening time scales with the number of molecules in the template:
1-molecule template = x time
2-molecule template = ~2x time
3-molecule template = ~6x time
4-molecule template = ~12x time
It’s definitely a smarter experiment to run each template molecule separately followed by a comparison of the results.
References
More information about Forge (previously known as FieldTemplater) and Blaze in the literature can be found in: (a) J. Med. Chem., 2008, 51, 565-573; (b) J. Comput.-Aided Mol. Des., 1995, 9, 33-43; (c) J. Med. Chem, 2005, 48, 6790-6802.

Rae Lawrence
Technical Sales North America