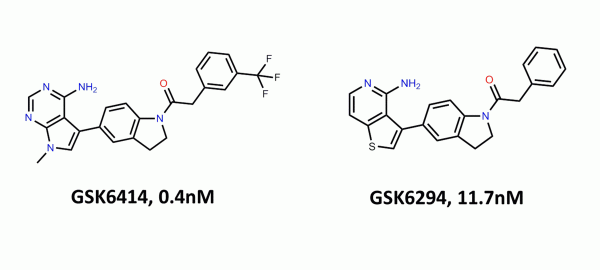
Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
After attending the ACS National Meeting in New Orleans and standing in ridiculously long lines for coffee, it is blatantly obvious that we chemists, as a group, have a serious addiction to coffee. It was quite interesting to see several tables of chemists, each person frantically scribbling structure ideas on their napkins, debating with their colleagues, looking intently at the structures before neatly folding the napkin and putting it in their pocket.
Each of these chemists was doing what chemists do best – designing the next set of compounds that would hopefully prove to be more favourable. “Favourable” could mean increased activity/potency, decreased toxicity, better ADME properties, or ideas to open up new intellectual property space.
Given this observation, this month, I’d like to share how Cresset’s software can be used as a “Virtual Napkin”. Like a paper napkin, structures would be scribbled down. But what makes Cresset’s tools different is that a chemist can get immediate feedback on how their newly designed compound (or modifications) stack up to a starting point compound.
Background for PERK Kinases
A recent publication1 from GSK’s Oncology Research unit reported the discovery of a series of potent and selective inhibitors of PKR-like endoplasmic reticulum kinase (PERK). Since I wasn’t really familiar with this kinase, I thought it would be interesting to see if we could take the structures and activity data within the paper to rationalize the structure-activity relationship with Torch and use this software as a “virtual napkin” to come up with modifications for an iterative design process.
Accumulation of unfolded proteins is becoming known as a factor in diseases such as neurodegeneration, heart disease, diabetes and cancer. This accumulation of unfolded proteins causes stress on the cell’s endoplasmic Reticula.2 To cope with this stress, the cells have an unfolded protein response (UPR) which causes a cell signaling cascade to alleviate the stress so that the cell may survive.3
It has been shown that several types of tumors demonstrate elevated levels of UPR-related proteins.4 In a number of papers cited in (1), protein kinase R (PKR)-like ER kinase (PERK) is a key factor in managing the UPR response. PERK is a type-1 ER membrane protein consisting of a domain facing the lumen of the ER which senses “stress”, a segment that is transmembrane, and a kinase domain facing the cytosol.
When PERK is inhibited in cancer cells, the viability of those cells in nutrient or oxygen deprived conditions is reduced, which can lead to apoptosis and inhibition of tumor growth. Until the GSK paper1, there were no known selective inhibitors of this target – the GSK compounds reported showed potency and selectivity against PERK, resulting in growth inhibition of humor tumor cells in mice.
Experiment
Before embarking on any in silico experiments, some examination of the protein-ligand crystal structure was required to answer the following questions:
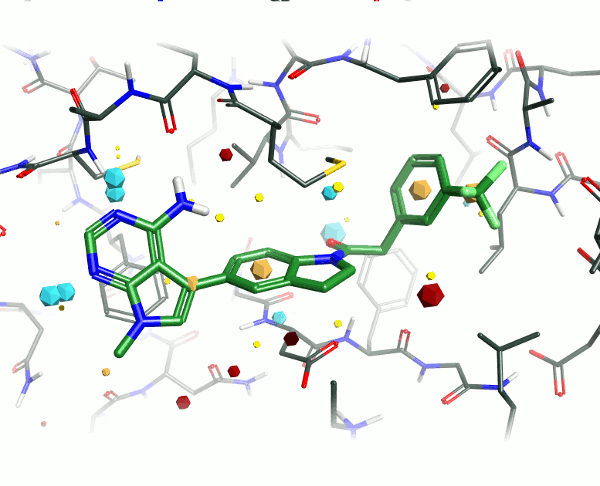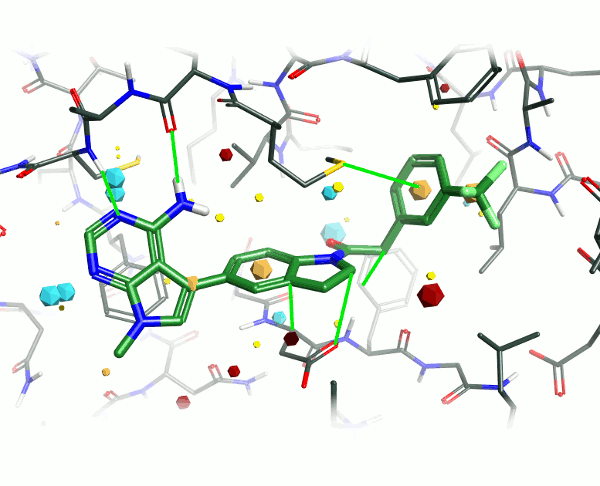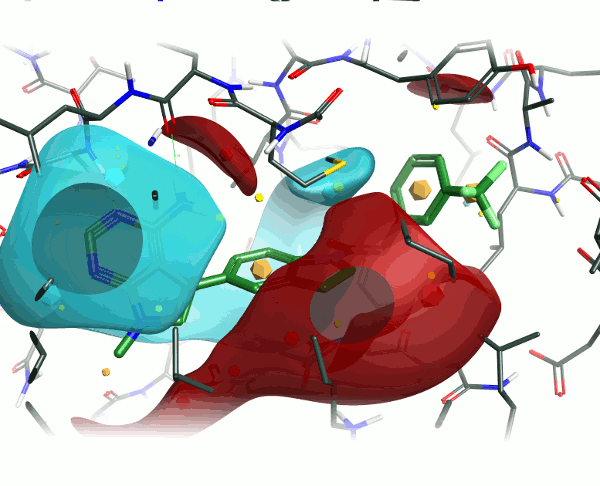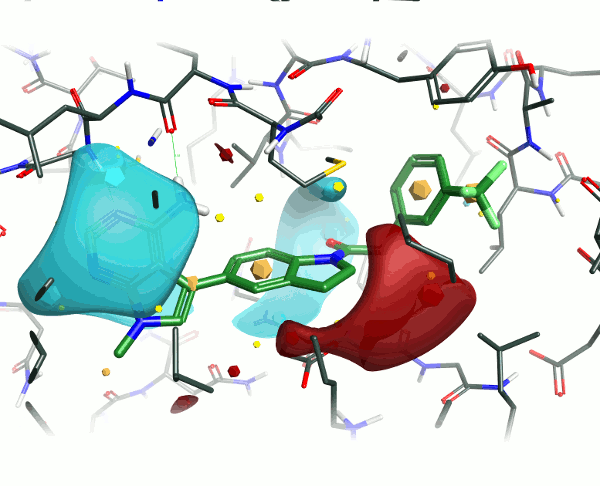
Looking at the 4G31 (GSK6414) ligand in the context of the PERK pocket, there are several key ligand-residue interactions important to the binding.
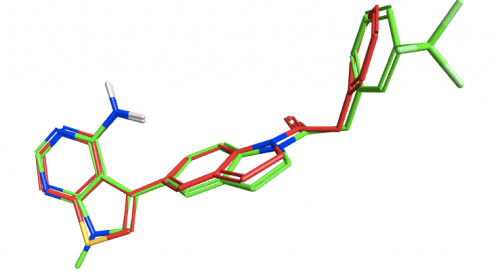
To examine the structure-activity relationship, the other GSK compounds from the article were brought into torchV10 as SMILES strings (copy-paste from ChemDraw, drawing with Torch’s built-in Molecule Editor are also structure input options), and aligned to the GSK6414 reference molecule using the “substructure” routine to ensure that the indoline core was retained in the alignment.
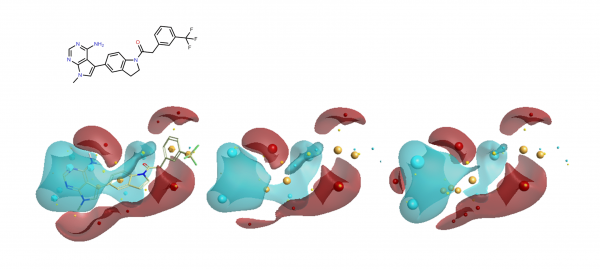
During this study the group looked at a number of different hinge binding groups. In this series the thienyl-pyrimidine and pyrrolo-pyrimidines were the most active (pIC50 9 vs pIC50 8 for the others) suggesting that the electronics of these groups are desirable. As noted in the paper, the activity differentiator for these systems is likely to be found in more subtle effects, since both size and shape requirements are all satisfied. The electrostatic field surfaces for six examples (figure 4) clearly show distinct differences across the set – whilst the thienyl and pyrrolo -pyrimidines are the most similar to each other.
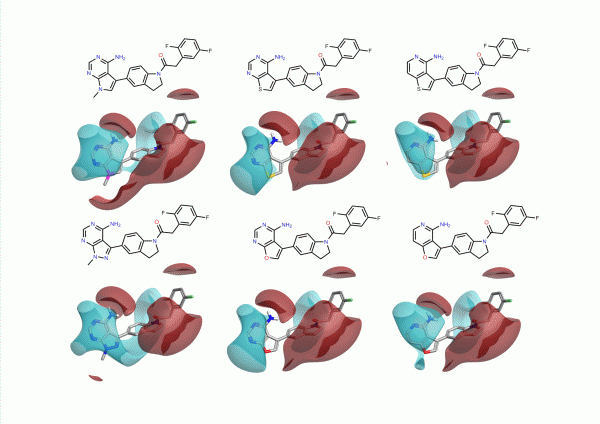
Classically medicinal chemists will think of many other heterocycles that could be used in this situation, often sketched on their napkin over lunch or on fumehood doors in the lab. Deciding which of these ideas to choose is not straight forward involving considerations of physicochemical properties, synthetic tractability, and intellectual property as well as activity. Cresset’s Torch is designed to help make these decisions easier by modeling the complex electrostatics of the system and presenting the results together with physicochemical properties in a simple interface.
We chose to design new heterocycles by moving nitrogen atoms around the 6,5 ring system. We didn’t consider synthetic utility on this occasion. The results are shown in Figure 5. Again we have modeled the difluorophenyl acetamide analogue to draw a direct comparison to Figure 2.
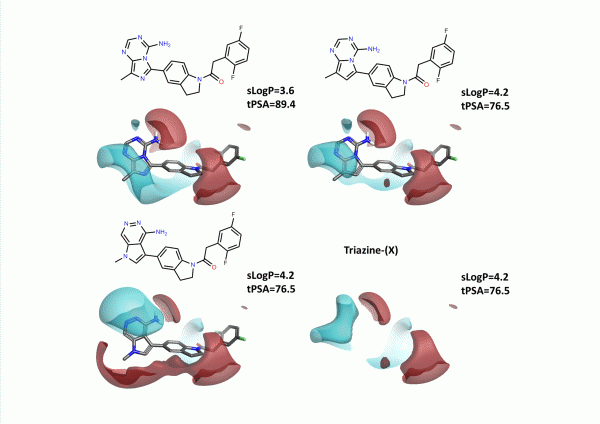
An alternative approach was to use Spark to find new heterocycles in this position. In this case we ran a Spark experiment using the initial CF3-substituted arylacetamide (GSK6414). Incorporating fragments from the common commercial database within Spark produced the 4 most potent example hinge binding heterocycles within the top 8 output compounds (figure 6), along with the triazine (X) example above. Interestingly the weaker thiopheno and furano – pyridines were not amongst the top ranking output – however a number of novel heterocycles with good electrostatic fit to the actives were retrieved which were not reported in the article.
References

Rae Lawrence
Technical Sales North America