Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
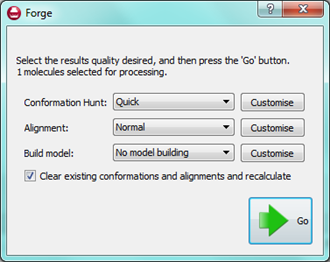
forgeV10.1 was released in March 2013 with a number of new features. We also made a number of improvements to usability and workflow in response to user feedback and our own internal project work. We use our software on an everyday basis for our consulting projects, so we know the difference that these improvements make to everyday tasks.
In this article I’ve highlighted how some of these new features can be put to work in projects. I’ll also go through a couple of tips and tricks that I’ve found useful when working with Forge myself.
One of the key tasks in Forge is to design new molecules and to edit the alignments of existing molecules. Since the molecular editor is used for both these tasks we have simplified how you tell Forge which task you are performing. This is done when you exit the editor – you choose either to save the alignment that you have just created by clicking the “OK” button or you choose to “Process” the molecule and have it aligned to the reference.
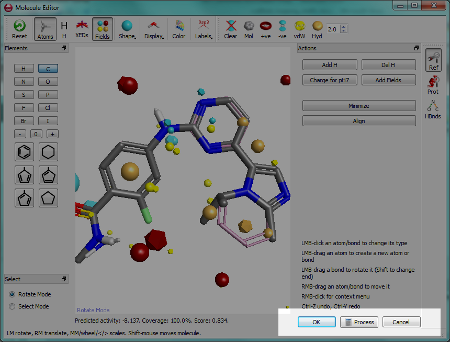
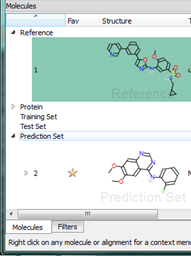
Right clicking on a molecule in the results table gives access to many commands in addition to those described above. For example if you have loaded a molecule file straight into Forge then it will not have been aligned. Using the right click menu you can now choose to promote any alignment of any molecule up to a reference molecule. Alternatively you can copy the alignment to the clipboard or delete it entirely.
The “Mark as aligned” entry can be used to convert the current coordinates of a molecule into an alignment, which is useful when you’ve read in structures that were aligned elsewhere, e.g., by protein sequence alignment. If you have a structure with conformations but no alignments (either from a file or by deleting alignments for a molecule) then the “Mark as aligned” right click menu entry will convert the conformations into alignments enabling a visual inspection of the population.
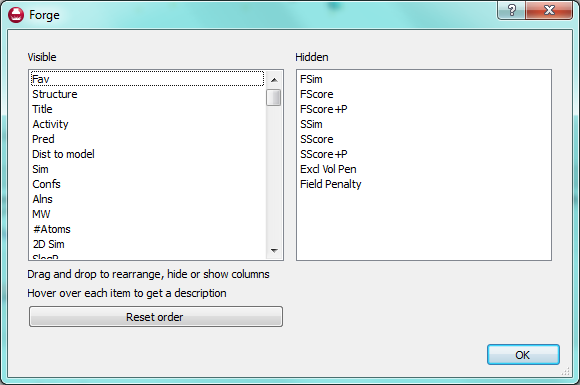
In Forge, molecules belong to “sets” that are named for QSAR. However, you can use these sets to separate out your dataset in whichever way works for you. For instance, you could load different chemical series into each of the sets or split molecules by activity into different sets. You can put molecules into a set at any time, for example when you find an interesting result you can simply drag and drop it into one of the other molecule sets (or use the right click menu to change its “role”) to have it saved separately. We are planning to enhance this feature in future versions to provide a customizable list of sets that you can rename, but we are keen to hear of other ways we can improve this feature.

Dr Tim Cheeseright,
Director of Products