Future-proofing Cybersecurity in Drug Discovery
The pharmaceutical and biotech sectors suffer more data security breaches than any other industry, with 53% resulting from malicious activity. To protect against potential ...
News
Many biological events are a consequence of the interaction of a cascade of proteins, each one triggering the next. Organisms can often work around one, or even two, missing or damaged components; but we suspect they struggle to complete processes that are impaired at three or more points [01].
The historical way of dealing with this situation was to administer several drugs in a cocktail, each of which hits an individual component. However, this practice is unsatisfactory on several levels.
The most effective solution would be a single drug that intervenes at multiple points of the protein cascade, acting with multiple functions and at multiple sites. This is what we understand as the definition of polypharmacology.
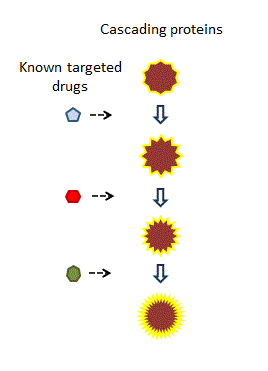
Historically, a cocktail of drugs may have been administered, each addressing a different component of the reaction.
Polypharmacology is recognised as a desirable approach to targeting disease conditions. In fact, it is rare to find a condition that requires only one set of characteristics from a drug compound. Taking a fields based approach to polypharmacology makes it possible to create a pharmacophoric template that combines the required characteristics. The following example illustrates this.
In the latter half of the last decade, Dr. Iain Chessell was involved in the search for a drug that overcomes neuropathic pain as opposed to nociceptive pain. He put forward a theory that neuropathic pain could be countered by blocking peripheral NaV1.7 sodium channels and α2-δ Ca channel subunits in the central nervous system [02]. This led to the requirement for a drug with not one, but three characteristics. The drug must inhibit NaV1.7, block the α2-δ Ca channel and in addition pass through the blood brain barrier (BBB).
In collaboration with Dr. Chessell, the Cresset team searched the literature to find drugs that matched each of the three requirements, resulting in two sets of compounds. Fortunately, the requirement to pass through the BBB was inherent in the drugs used to block a α2-δ so the two sets were fit for purpose. At this stage one possibility would have been to recommend a cocktail of one drug from each set. Instead, we continued the search for one drug that would contain the properties of all three targets.
Cresset’s technology allows us to define the action for a drug not through its structure, but through its field pattern. If we can define three field patterns for each target, they can be combined to form a single compound field pattern. We can then search our databases for a molecule with a similar pattern.
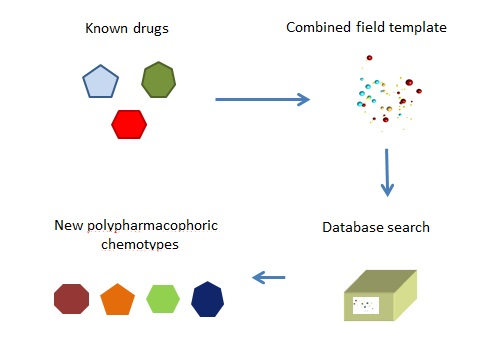
This is used to search for a compound that combines the required characteristics.
For our neuropathic pain project, the literature search had yielded enough compounds from each set to generate a reasonable pharmacophore template based on the field patterns.
A field template for NaV1.7 inhibition was generated by finding a common pattern across tetrodoxin, saxitoxin, Xenon 1.7 inhibitors, MSD M9, lamotrigene and carbazepine.
A field template for blocking the α2-δ Ca channel and overcoming the BBB was derived from pregabalin, gabapentin plus six variations and four 5-hetero ring derivatives.
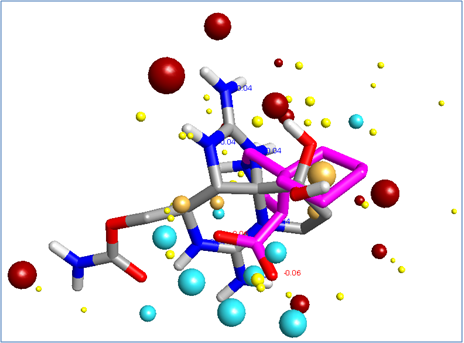
Overlaying these field-based pharmacophores resulted in a polypharmacophore that was used to search a database of compounds (Figure 3).

This process yielded a number of promising results that are yet to be tested. The bioactive conformations of Saxitoxin (grey) and Gabapentin (pink) superimposed in 3D into the same field template are illustrated in Figure 3, below. The molecules both fit into the polypharmacophore very well, but the smaller gabapentin (pink) structure is satisfying field points towards the right of the pattern below, which represents the section of the polypharmacology particularly important for α2-δ activity. The larger Saxitoxin molecule satisfies more of the field points towards the left hand side of the pattern below, which are associated with NaV1.7 activity.
This same approach can be applied to searching for compounds that hit multiple proteins in a cascade. Polypharmacophores can be created for different combinations of the proteins. For example, in a cascade of 8 proteins, different combinations of three to four proteins can be used to create a series of polypharmacophores. These can be used to search for compounds that will hit different combinations of proteins.
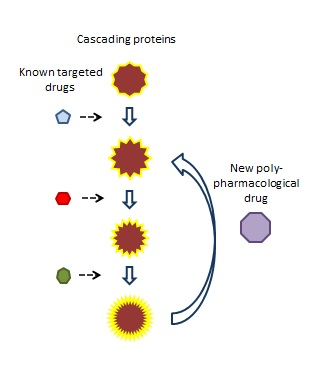
It is interesting to consider how this approach can also be applied to selectivity through the use of negative field patterns.
Selectivity and polypharmacology can be seen as two sides of the same coin. Whether it is activity or inactivity, the goal is to combine a set of ideal characteristics for interactions at different targets.
Chemical libraries are often built based on a central scaffold with added chemistry. However, this does not result in a diverse library and the chances of activity at the required sites are low.
An alternative approach to building a diverse library is as follows.
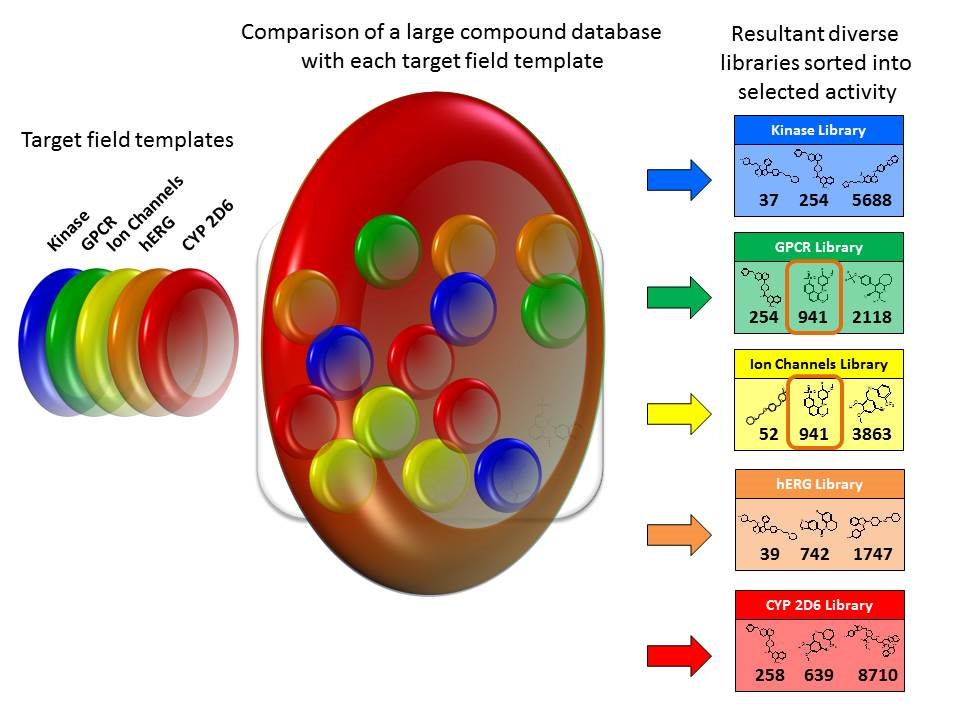
While some of the approaches outlined above are speculative, enough work has been done on creating polypharmacophores to demonstrate the viability of fields based polypharmacology. As computing power continues to increase, so too will the scope of fields based searching for polypharmacophores.