Future-proofing Cybersecurity in Drug Discovery
The pharmaceutical and biotech sectors suffer more data security breaches than any other industry, with 53% resulting from malicious activity. To protect against potential ...
News
One of the most common questions we get asked at Cresset relates to our handling of conformations. Our alignment technique uses rigid-body rotations, so we rely on computing conformer populations for molecules whose bioactive conformation is unknown. The generation of useful conformers of flexible molecules is old science: programs have been around since the 1970s tackling it in one fashion or another but it remains an active challenge nearly 50 years later.
The conformer analysis problem can be broken down into two parts: what does the conformational energy landscape look like, and where are the minima in that landscape? The difference between conformer searching techniques largely falls into the latter part: global optimization problems are inherently hard, especially in high-dimensional spaces, and so there are a plethora of different ways of handling them: systematic search, Monte Carlo techniques, simulated annealing, low mode molecular dynamics, divide-and-conquer methods, rule-based expert systems, genetic algorithms, and so forth. These days there is a wide variety of free and commercial tools using one or more of these methods to choose from, all of which (of course) claim to be the best.
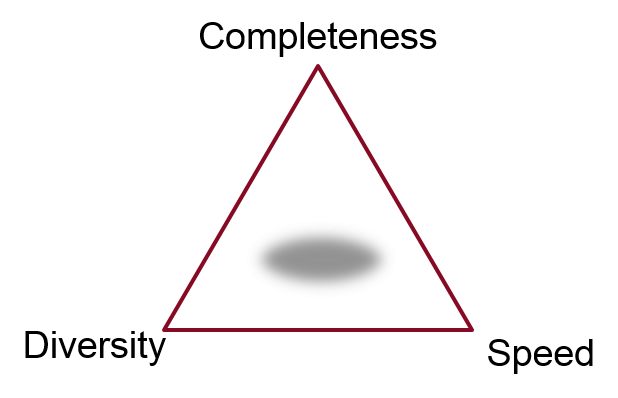 What do we mean by ‘best’, though? The answer, as with so many things in science, is it depends what you want to use the output for. An appropriate technique for doing an exhaustive enumeration of the conformer space for cyclononadecane may be completely useless if applied to the problem of generating a small set of conformations as input for a 3D clustering problem. There’s generally a three-way tradeoff: completeness, diversity, and speed. Do you need all of the possible conformers, or just a representative sample, and how long are you willing to wait?
What do we mean by ‘best’, though? The answer, as with so many things in science, is it depends what you want to use the output for. An appropriate technique for doing an exhaustive enumeration of the conformer space for cyclononadecane may be completely useless if applied to the problem of generating a small set of conformations as input for a 3D clustering problem. There’s generally a three-way tradeoff: completeness, diversity, and speed. Do you need all of the possible conformers, or just a representative sample, and how long are you willing to wait?
 If you need to compute thermodynamic properties of a molecule using a Boltzmann average across its conformer space, then complete searching of that space is essential. However, if you’re doing a 3D virtual screen across 107 molecules, then fully exploring conformation space is not necessarily a benefit. You are more likely to find the correct conformer for the actives that will give them a high score, but you’re also more likely to find a conformer for the inactives that also results in a high score. So the alignments and scores get better, but the enrichment does not. Accordingly, an exhaustive conformer search for virtual screens is usually both unnecessary and prohibitively costly, and the important question is how well you can cover conformational space in a limited number of conformers and minimal CPU time.
If you need to compute thermodynamic properties of a molecule using a Boltzmann average across its conformer space, then complete searching of that space is essential. However, if you’re doing a 3D virtual screen across 107 molecules, then fully exploring conformation space is not necessarily a benefit. You are more likely to find the correct conformer for the actives that will give them a high score, but you’re also more likely to find a conformer for the inactives that also results in a high score. So the alignments and scores get better, but the enrichment does not. Accordingly, an exhaustive conformer search for virtual screens is usually both unnecessary and prohibitively costly, and the important question is how well you can cover conformational space in a limited number of conformers and minimal CPU time.
The first part of the conformer analysis problem (what does the energy landscape look like?) is often overlooked, but is actually the most interesting bit. In drug discovery, we’re most interested in bioactive conformations i.e. the shape of the molecule when bound to the protein active site. So, what conformational energy is appropriate? In vacuo energies are clearly not very helpful, as intramolecular van der Waals and electrostatic interactions are inappropriately emphasized. In particular, large-basis-set ab initio calculations with no solvation model can be a good way to ascertain local conformational preferences of particular functional groups but are of very limited use on flexible drug-sized molecules.
So, if vacuum calculations are no use, should we just use a solvation model of some sort (GBSA, PB, or whatever)? Well, yes. And no. The problem is that the interior of a protein doesn’t look much like water, so the most favorable conformer inside an active site can be quite a long way away from the most stable solution conformation: estimates of how far off vary, but reports in the literature suggest that values of at least 3-5kcal/mol (calculated using force fields)1,2 to more than 25kcal/mol (from large ab initio calculations with continuum solvation model)3 are needed if you to keep the bioactive conformation. Given this range, is the ‘lowest-energy conformer meaningful?
These energy gaps are partly due to the effect of protein-ligand fitting, and partly (in the molecular mechanics case) due to inherent errors in current force fields. The XED force field is one of the best available at reproducing experimentally-determined conformation energies, but even it has errors of ca. 0.3-0.4 kcal/mol on its standard validation data set. Since the experimental data tend to be on very small molecules, the errors on conformer energies of drug-sized molecules will be more like 2kcal/mol from this source, and the inaccuracy of continuum solvation models will increase this further. The fact that the computed strain energy for bioactive conformations increases with the flexibility of the molecule bears this out.1
 Some conformer exploration programs avoid the messiness of force fields by using heuristics and small molecule crystal data to generate conformers. The CSD is a wonderful source of experimental data, and aggregate statistics of torsion angles etc. can be very useful. However, there can be a tendency to assume that individual CSD-derived conformers are inherently better/more useful than others, and are somehow closer to the “truth”. However, many structures in the CSD are heavily affected by crystal packing forces: the structure CUCMES, for example, has an axially-substituted cyclohexane, while POVSEY has a diaxial 1,4-dihydroxycyclohexane. Statistical torsional profiles are great, but pulling an individual fragment from a CSD molecule and claiming that it is a “better” conformation because it is from experimental data is misguided.
Some conformer exploration programs avoid the messiness of force fields by using heuristics and small molecule crystal data to generate conformers. The CSD is a wonderful source of experimental data, and aggregate statistics of torsion angles etc. can be very useful. However, there can be a tendency to assume that individual CSD-derived conformers are inherently better/more useful than others, and are somehow closer to the “truth”. However, many structures in the CSD are heavily affected by crystal packing forces: the structure CUCMES, for example, has an axially-substituted cyclohexane, while POVSEY has a diaxial 1,4-dihydroxycyclohexane. Statistical torsional profiles are great, but pulling an individual fragment from a CSD molecule and claiming that it is a “better” conformation because it is from experimental data is misguided.
So, having avoided all of these myths, what should you do? The answer of course depends on what you want to use the conformers for. In our case, we want conformations to maximize either (a) the chances of finding the correct alignment of a ligand to a reference structure, or (b) the enrichment from a 3D virtual screen. In both cases, the trade-off between coverage and CPU time dictates that we want to generate a diverse set of reasonable conformations rather than a full systematic search, but in the latter case we can generally get away with somewhat fewer conformations on average. Either way, we treat all low-strain conformations on an equal footing.