Future-proofing Cybersecurity in Drug Discovery
The pharmaceutical and biotech sectors suffer more data security breaches than any other industry, with 53% resulting from malicious activity. To protect against potential ...
News
The XED molecular mechanics force field has been used to analyze the binding between β2AR and a G-protein and the difference in binding speed in the presence of agonists and antagonists.
The challenge was to model the complete structure of the entire system at every stage of a process that lasts in reality maybe milliseconds, to find the energetic minima at each stage.
An adjustable length constraint was applied to the position of the receptor relative to the G-protein so that the energies could be calculated for a series of positions as the receptor separated from the G-protein in the presence of an agonist or an antagonist.
Thus computational calculations were made of the energetics ‘along a line’ using the Cresset FF3 force field, based on experimental data. These calculations were used to carry out an analysis of the stability of the resultant complex in terms of the G-protein binding energy.
Aromatic-aromatic interactions are vitally important to protein structure, and the XED force field is particularly suited to modelling these types of interaction.
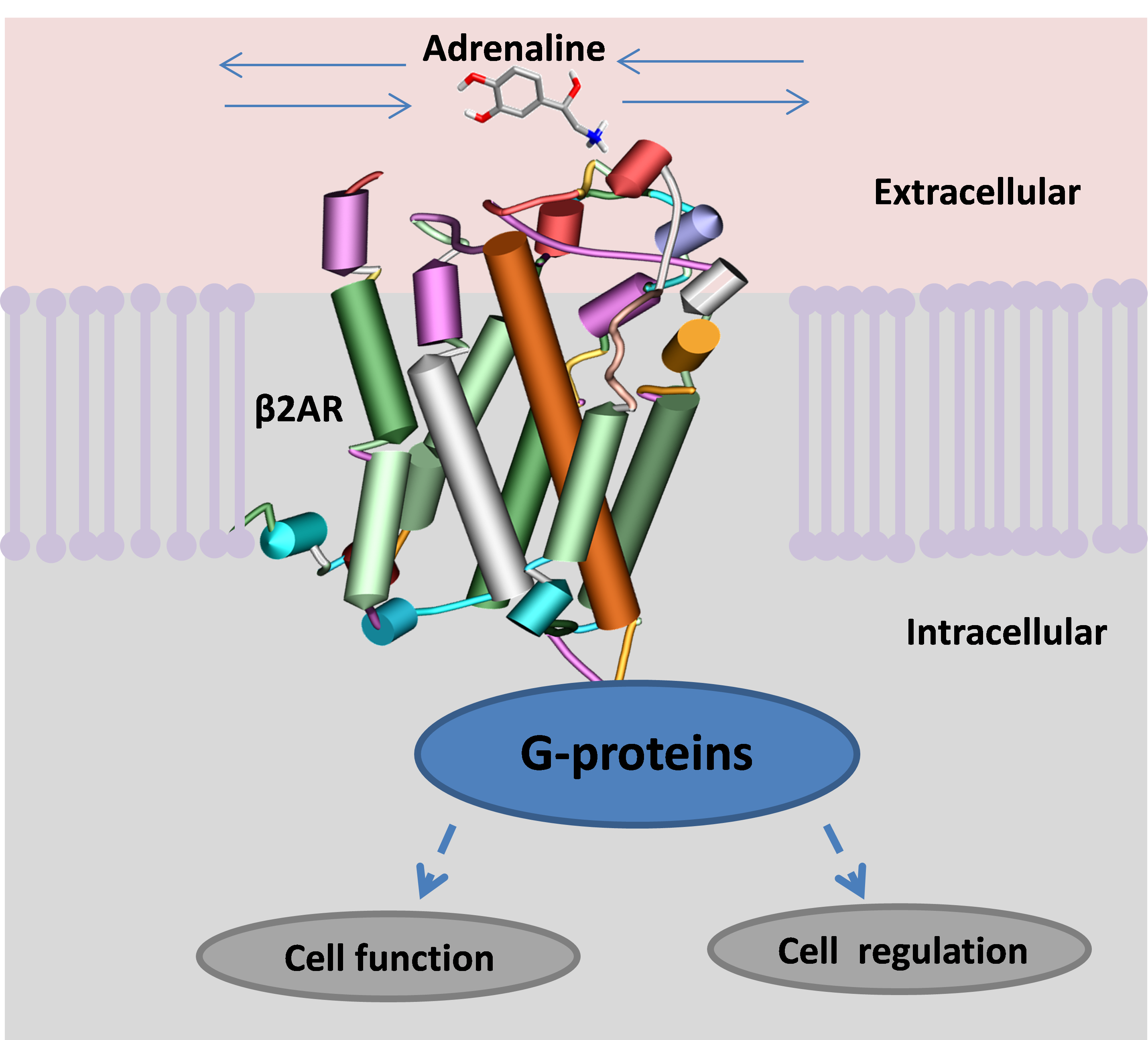
For thirty years, I worked with Sir James Black, the inventor of propranolol; the first drug to counter high blood pressure. This drug acts on the β2-adrenergic receptor (β2AR), one of the receptors that controls heart function. β2AR is a member of possibly the most ubiquitous and important receptor family in the human body; the G-protein coupled receptors (GPCRs). About 40% of all available drugs act on GPCRs.
A GPCR spans the membrane of a cell. When activated it binds with a G-protein in the cell, which starts several vital processes, the most important of which is the creation of fuel for the cell’s engines. The β2AR is activated by recognising a small molecule, adrenaline or noradrenaline, from the blood stream and binding it into a specific active site. Once ensconced, the small ligand changes the chemistry across the receptor, allowing it to bind to a specific G-protein on the inside of the cell that switches on a series of actions (figure 1).
Many drugs exist that simulate adrenaline actions and are termed receptor ‘agonists’
Propranolol, discovered by James Black, was the first example of a β2AR ‘antagonist’; a drug that blocks the ability of the receptor to bind its G-protein, effectively deactivating the receptor. Propranolol has the effect of controlling an overactive heart, and thus reducing blood pressure. Many synthetic agonists and antagonists have appeared since propranolol and continue to save millions of lives across the world.
However, the molecular mechanism by which these ligands influence the receptor’s ability to bind its G protein remains confused and indefinite.
It is important to point out that the β2AR is known to undergo spontaneous activation in the absence of any ligand. This is known as constitutive agonism and is some 200 times slower than the rate of normal agonist-induced activation.
Experimental data on the structure of the β2AR became available in 2009. In 2011 several X-ray crystal β2AR structures were published incorporating different ligands in the active site, including both agonists and antagonists.
The puzzling thing was that, even though the X-ray structures had both agonists and antagonists in their active sites, there were no helpful differences in the X-ray structures. Figure 2 shows six overlaid X-ray structures, four of which contain antagonists and two containing agonists. There is no discernable difference in structure.
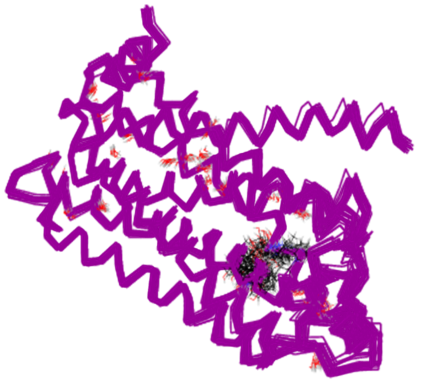
The expectation had been to find the protein in an active (R*) state when bound to an agonist, showing an interpretable conformational change from the inactive (R) state when bound to an antagonist. This led to a great deal of speculation in the literature to try to reconcile the results with theory, some of which was nothing short of wishful thinking!
I started to try to get some feeling for whether the force field gave reasonable answers for the binding of the ligands. It was a lot of work and the results weren’t provable. Things were consistent, but computing work is always speculative. There was no real experimental evidence for what we were seeing and calculating.
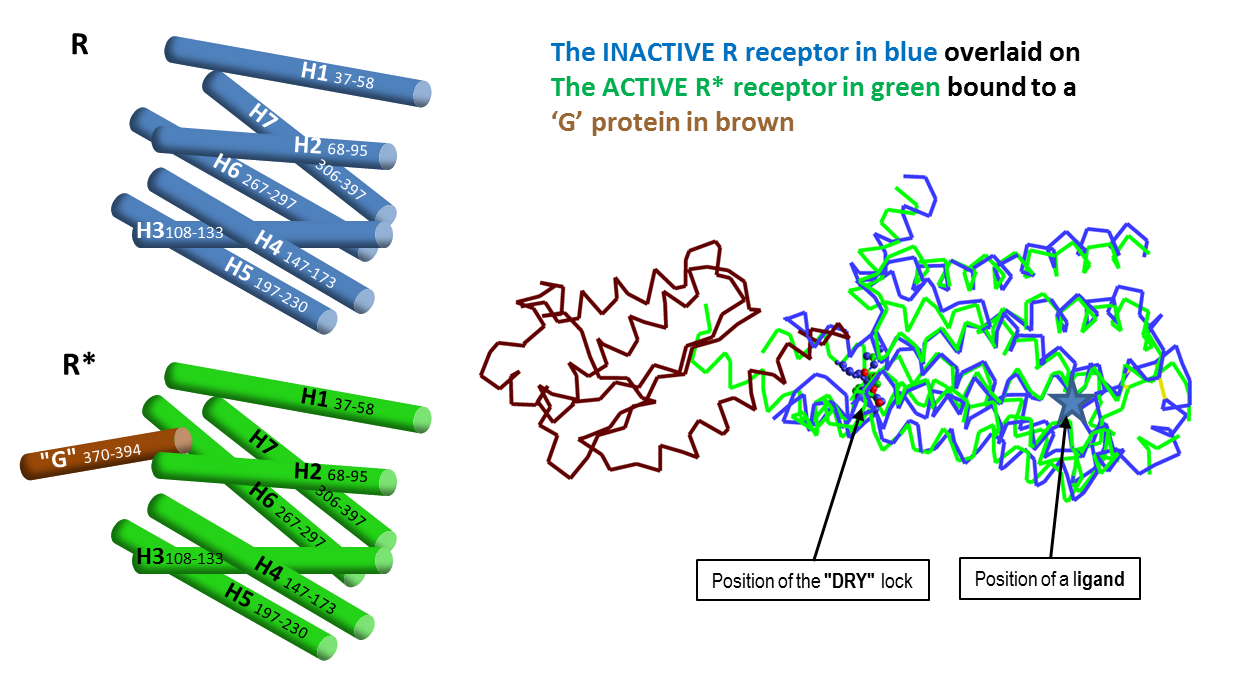
In July 2011 everything changed. Brian Kobilka published an X-ray structure of the activated β2AR connected to a G-protein surrogate. This was the first X-ray structure of any GPCR in its active state actually turning on the G- protein. It was in a totally different form from all of the x-rays that had been done before. Kobilka had found the active R* β2AR.
In Kobilka’s words: “A comparison of the inactive structure and an antibody stabilised active structure shows how binding events at both the extracellular and intracellular surfaces are required to stabilise an active conformation of the receptor. …that an agonist-bound active conformation spontaneously relaxes to an inactive-like conformation in the absence of a G protein or stabilising antibody.” Kobilka et al.2011 Nature 469, 175. Kobilka et al, ibid. 236. Warne et al. ibid. 241.
The conclusion was that the activated R* form of the GPCR never forms unless it’s linked to its G protein, whether or not an agonising or antagonising ligand is present.
The Nobel Prize in Chemistry 2012 was awarded jointly to Robert Lefkowitz and Brian Kobilka for ‘studies of G-protein-coupled receptors’.
At this stage, I proposed that the ligands were changing the ease with which the G-protein was allowed to enter and exit the β2AR G-protein binding site. You can imagine that the active form wants to hold the G-protein, whilst the inactive form wants to get rid of it. Antagonists discourage the GPCR from picking up the G-protein whilst agonists encourage it. I reported this proposal in my lecture at the MGMS Downing meeting in Cambridge in the summer of 2011.