Future-proofing Cybersecurity in Drug Discovery
The pharmaceutical and biotech sectors suffer more data security breaches than any other industry, with 53% resulting from malicious activity. To protect against potential ...
News
Cresset’s powerful scaffold hopping and fragment replacement software can be used to mimic an existing ‘active’ molecule’s electrostatic patterns to quickly, and efficiently, generate new molecular designs. The outcome, based on 3D molecular electrostatic similarity, is more biologically relevant than that from other similarity metrics. Here, we show how Spark can be used to rapidly generate potentially valuable chemical ideas for new DPP-IV inhibitors.
Two peptide hormones, GLP-1 and GIP, mediate lowering of blood glucose level through stimulation of insulin release and inhibition of Glucagon release. DPP-IV cleaves a dipeptide from the N-terminus of both GLP-1 and GIP hormones to give their inactive forms, thus abolishing their glucose lowering action. DPP-IV inhibitors have been shown to be important agents useful for treating type II diabetes.
The method involves:
A plethora of molecules are already known which inhibit DPP-IV and have useful anti-diabetic properties.
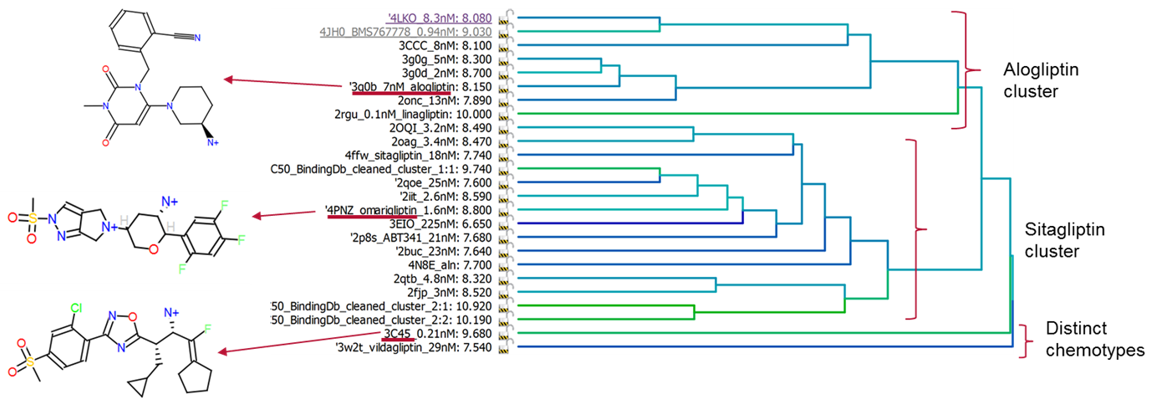
Superimposition of over 20 published x-ray crystal structures, followed by hierarchical clustering using all-by-all field similarity on the ligands, reveals two main clusters and a distinct mode of binding for a fluoro-olefin example (Figure 1). This unique clustering, performed in Cresset’s ligand-focused workbench, Forge, allowed the selection of three distinct inhibitors for further scaffold hopping work.
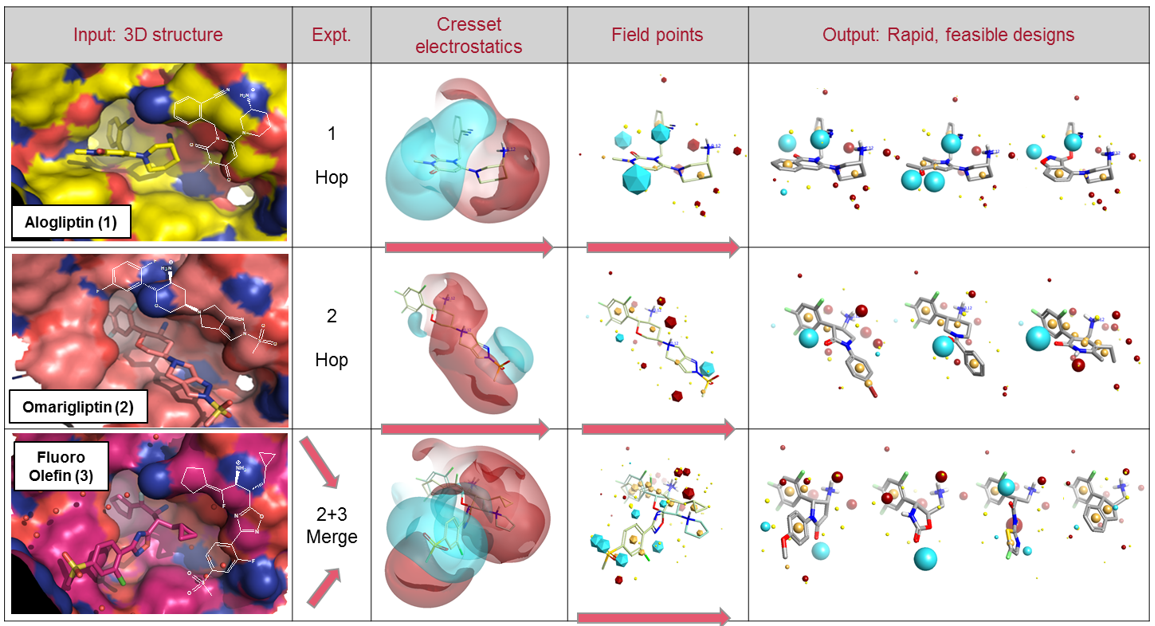
Alogliptin (1), Omarigliptin (2) and the fluoro-olefin (3, PDB: 3C45) represent some of the most ligand efficient examples from these clusters. Two experiments were performed in Spark using (1) and (2) in a simple scaffold hopping exercise. A final experiment was a chemotype merging experiment: a truncated (2) was used, with (3) as a second template, to find molecules bridging the two series. Workflow as shown in Figure 2.
Example results (Table 1) shows the diverse range of output suggestions provided for new chemistry and validates the method by providing examples which already have precedents in patents and the literature.
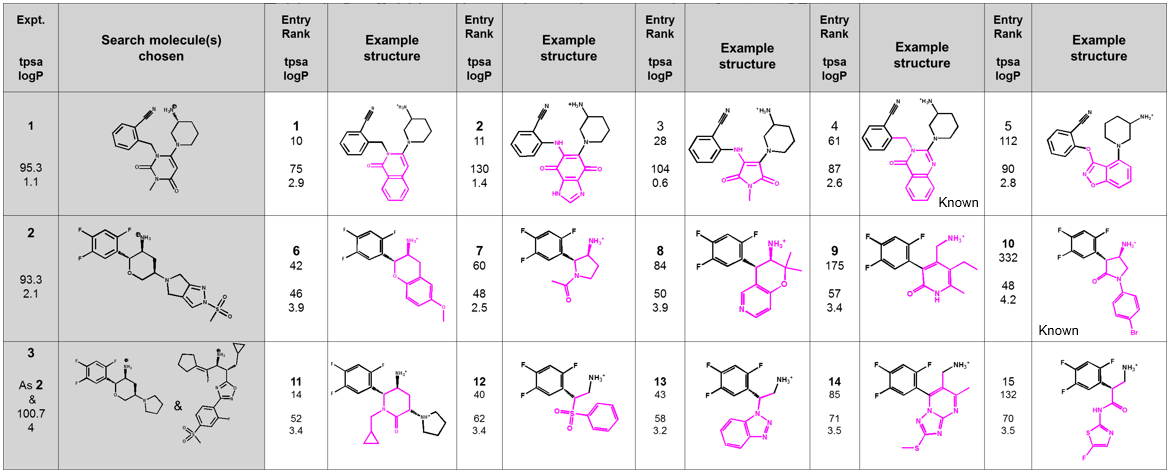
The Spark searches output a wide range of diverse chemotypes that included active or very close architectures to known active frameworks. A number of these had been discovered through HTS rather than through rational design. Tight control over the chemistry ensures that feasible chemistry is provided from known fragments whilst maintaining the features necessary for activity.
Spark is a powerful molecular modeling tool for the rapid virtual elaboration of scaffold ideas; either in scaffold hopping, merging, fragment growing or linking experiments. Applying Spark to chemotypes bound to the active site of DPP-IV provided a range of interesting and synthetically-feasible suggestions.
Green et al, Diabetes and Vascular Disease Research 2006, 3 No3, p159-65.
Protein pictures were rendered using open source Pymol from Delano Scientific.
[This content was presented as a poster at Proteinase 2015.]