Future-proofing Cybersecurity in Drug Discovery
The pharmaceutical and biotech sectors suffer more data security breaches than any other industry, with 53% resulting from malicious activity. To protect against potential ...
News
Working with molecules in 3D is computationally expensive compared to most 2D methods. Most modern cheminformatics toolkits can do hundreds of thousands to millions of 2D fingerprint comparisons per second, with 3D similarity techniques being multiple orders of magnitude slower.
The computationally-expensive part of the 3D calculations usually involves aligning conformations to each other. The natural tendency therefore, is to see if we can skip this step and compute a set of properties that can tell us if two molecules are similar in 3D without actually having to align them. If this works we can get the best of both worlds: the speed of 2D comparisons combined with the accuracy and structural independence of 3D similarity functions.
The earliest version of this idea is the simple pharmacophore. All you have to do is assign a few pharmacophoric points to each molecule (usually based on some sort of functional group pattern recognition), then generate a set of descriptors based on sets of these (usually 2 or 3). If two molecules share one or more pharmacophore descriptor, then they match.
Pharmacophore searches succeed on some counts: they are indeed very fast, and they do encode some 3D information. However, they involve a very crude binning of the wide array of possible intermolecular interactions into a few pharmacophore types, and they describe shape poorly giving them very limited predictive power.
If pharmacophores can’t describe shape well, are there other techniques that can? A number of different methods have been presented, such as those based on multipole moments or spherical harmonic coefficients (e.g. ParaSurf/ParaFit), as well as methods based on statistical moments such as Ultrafast Shape Recognition (USR). None of these has achieved widespread use: harmonic coefficients are not rotation-invariant, while the USR technique correlates poorly to more accurate measures of shape similarity. 1
It would be nice if there was a way of providing alignment-independent descriptors which described both electrostatics and shape/pharmacophoric properties with a reasonable degree of accuracy. This is actually one of the first things we did when we were looking into starting Cresset – we developed a method called FieldPrint that encodes the distance matrix of field descriptors down into a fingerprint that can be used for alignment-independent similarity calculations. The concept is similar to that of GRIND 2 which was published around the same time, although the algorithmic details are somewhat different.
We put a lot of work into these techniques, but were never able to get a method that we were completely satisfied with. The problem we found is that encoding the distances in pairs/triplets of field descriptors ends up losing too much 3D information, and as a result you either end up with a slower mimic of standard 2D fingerprints, or you end up with a large false positive count. The FieldPrints have a tendency to find molecules with a similar overall pattern of positive/negative field, but can compute a very high similarity for molecules that are in reality quite dissimilar in terms of the 3D spatial arrangement of those fields. My belief now is that this is an inherent flaw of alignment-independent descriptors: they either have to be sufficiently complex that you are in effect computing an alignment, or you lose too much information and are not significantly better off than just using old-fashioned structural/pharmacophoric fingerprints.
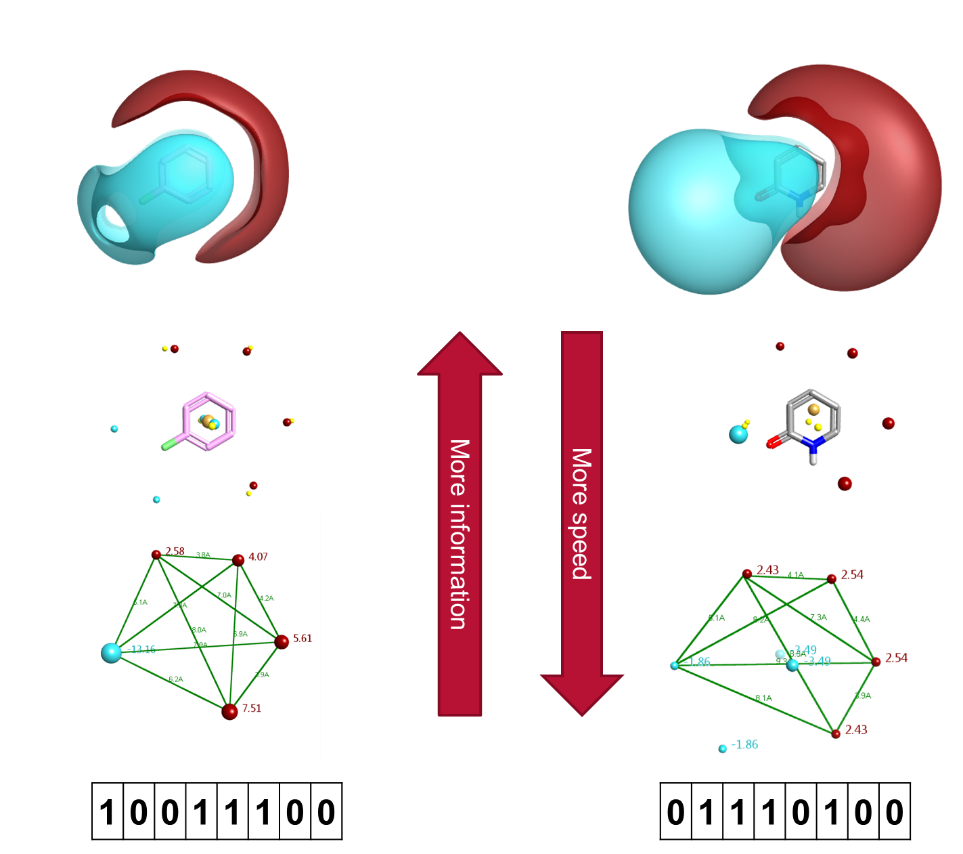
A further consideration is how you handle conformation. The original GRIND papers just use a single conformer per molecule, and their validation was confined to series of rigid molecules or sets of molecules where single conformations were generated and manually adjusted to be similar. In the general case neither of these shortcuts will work. Any method that purports to be 3D but starts with a single conformation per molecule is inherently flawed: the whole point of 3D is that molecules are flexible.
There is a disturbing number of papers out there that do some sort of notionally-3D analysis on set of single CORINA-derived conformations. You can get very good enrichment factors on retrospective virtual screens doing this, but in practise the enrichments are largely bogus. CORINA is deterministic, and as a result molecules with similar structures will tend to be put into similar conformations. Combine this with the fact that many standard retrospective VS data sets have very low structural diversity, and the problem becomes apparent. The query molecule and its dozens of congeners in the “actives” data set are all placed in the same single conformation, and so application of a 3D or pseudo-3D technique can easily produce excellent-looking enrichment statistics. However, the enrichment all comes from a hidden 2D similarity.
So, single-conformation methods are a dead end and we need to consider flexibility. Once you are doing so, you need to factor in both the conformer generation time as part of the build time for the descriptor, and also factor in that your comparison speeds will now be two to four orders of magnitude slower than 2D fingerprints (assuming 100 conformers per molecule, and depending on whether you know a single bioactive conformation for one of the two molecules being compared or whether you need to compare conformer populations). 2D methods thus have an unassailable speed advantage, which is part of the reason they remain so popular.
Our original vision for Blaze (or FieldScreen, as it was then) was that it would rely on the FieldPrints to give extremely rapid searching. You can get quite good enrichment factors from the FieldPrints in retrospective virtual screens, but when we investigated further this is largely because they act as a proxy for overall molecular size and charge. Once you control for that by more careful selection of decoys the FieldPrint performance is much less good. Analysing a molecular similarity technique through retrospective virtual screening performance is very very hard to do well, and as a result I am intrinsically wary of methods that present a set of DUD enrichments as their sole validation: FieldPrints perform quite well on DUD, but we know that they are not particularly effective in real prospective applications.
We still use the FieldPrint technology: it’s the first search stage in every Blaze run. It’s generally good enough to filter out 25-50% of decoy molecules that have no similarity to the query, but certainly not good enough to use the FieldPrint ranks directly. This is why we just use them as a pre-filter: molecules that pass that filter have much more accurate similarities computed using our alignment-based clique/simplex algorithms.
In the end, there’s no real short cut. All attempts to date to make 3D comparisons faster by simplifying descriptors and skipping the expensive alignment step just seem to leave out too much information – such techniques can be useful for cutting down the search space but if you’re going to spend CPU cycles working in 3D you might as well do it properly!
1. T. Zhou et al. / Journal of Molecular Graphics and Modelling 29 (2010) 443–449
2. Pastor, M.; Cruciani, G.; McLay, I.; Pickett, S.; Clementi, S. J. Med. Chem. 2000, 43 (17), 3233–3243.
Try our software for yourself – download a free evaluation.