Future-proofing Cybersecurity in Drug Discovery
The pharmaceutical and biotech sectors suffer more data security breaches than any other industry, with 53% resulting from malicious activity. To protect against potential ...
News
 Anupriya Kumar, Assistant Professor, Nagoya University, summarizes the publication Development of Small-Molecule Cryptochrome Stabilizer Derivatives as Modulators of the Circadian Clock which cites how Cresset software was used to understand SAR of circadian rhythm modulators.
Anupriya Kumar, Assistant Professor, Nagoya University, summarizes the publication Development of Small-Molecule Cryptochrome Stabilizer Derivatives as Modulators of the Circadian Clock which cites how Cresset software was used to understand SAR of circadian rhythm modulators.
Mammalian biological clocks control almost every physiological process such as metabolism, hormone secretion, body temperature and the sleep/wake cycle. Each cell in our body follows a circadian rhythm of 24 hours. Our modern lifestyle can disrupt this rhythm, for example working night shifts, or flying long distance. This in turn can lead to sleep and metabolic disorders such as obesity and type 2 diabetes. The daily oscillations of the circadian rhythm are generated by a loop which consists of clock genes such as CLOCK and BMAL1 (transcriptional activators) and two repressors: Cryptochrome (CRY1 and CRY2) and Period (PER1 and PER2).
The circadian rhythm can be modulated by small molecules, for example carbazole derivatives. One such molecule, KL001 was discovered by screening 60000 structurally diverse compounds in a cell-based Bmal1-dLuc reporter assay 1. KL001 lengthens the circadian rhythm by nearly 6 hours by stabilizing the clock protein cryptochrome (CRY). In the co-crystal structure of KL001 with the CRY2 protein (PDB ID. 4MLP 2), KL001 displaces the naturally binding FAD ligand (flavin adenine dinucleotide) while occupying only a small part of the binding pocket. Subsequent derivatization of KL001 resulted in a ten times more potent compound, KL044.
To understand the factors responsible for binding and stabilizing the CRY protein, we used Forge to perform 3D modeling and multivariate statistical analysis on 60 derivatives of KL001 and KL044. For generating a pharmacophore based on KL001 and KL044, we used FieldTemplater, which could reproduce the binding pose of KL001 to the CRY protein as shown in the crystal structure. We established a robust 3D-QSAR model by aligning all 60 compounds to the pharmacophore. The molecules were randomly divided into a training set of 54 and a test set of 6 compounds. Using this model, we successfully predicted activities of 5 new compounds and explained the evolution of the SAR by analyzing the contributing field points. By overlaying the field points from the 3D-QSAR model with the protein, we deciphered the important ligand-protein interactions.
Activity Miner was used to study differences in the activity of the compounds by analyzing the electrostatic potential. The differences in the electrostatic potential maps (red = positive potential, cyan = negative potential) of the two matched pairs are shown below. For example, introducing a disubstituted phenyl ring and an amide group (a) enhances the activity as does the presence of a mesyl group in the same position (b).
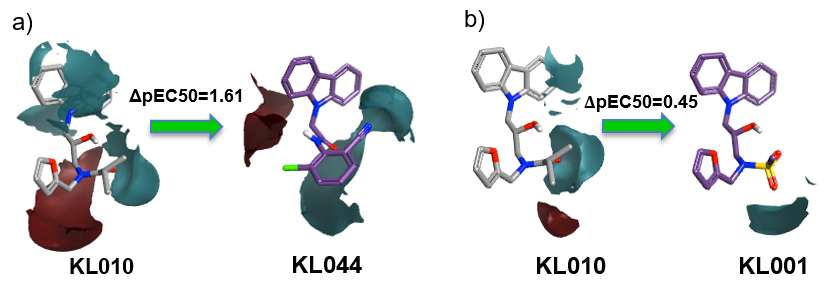
The current study serves as an encouragement for interdisciplinary teams to collaborate by showing that theoretical hypothesis and experimental studies complement each other. Knowledge based functional modifications can be made to KL001 and KL044 based on the activity enhancing key field points obtained from the 3D-QSAR model. Based on these findings, virtual screenings of compound databases can be performed to find novel compounds which would help in forwarding the field of drug discovery for circadian rhythm modulators which could lead to potential drugs for diabetes and other metabolic disorders.
1. Identification of small molecule activators of cryptochrome. Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T, Pongsawakul PY, Sonntag T, Welsh DK, Brenner DA, Doyle FJ 3rd, Schultz PG, Kay SA. Science. 2012 Aug 31;337(6098):1094-7.
2. Nangle S, Xing W, Zheng N. Cell Res. 2013 Dec;23(12):1417-9.