I am delighted to announce the release of a new version of Spark™, our scaffold hopping and bioisostere replacement tool. The focus of V10.5 is on advanced workflows and improved database management but also includes new science and many new and improved features.
The most interesting new features are presented below, and I encourage you to try this new release for yourself to see them in action.
Highlights
- New wizards to support ligand growing and linking, macrocyclization and water replacement experiments
- Enhanced Spark database update functionality
- New pharmacophore constraints
- Enhancements in search algorithm and advanced options.
New Spark wizards
The new Spark wizards will help you set-up advanced bioisostere replacements experiments in a user friendly and scientifically robust manner.
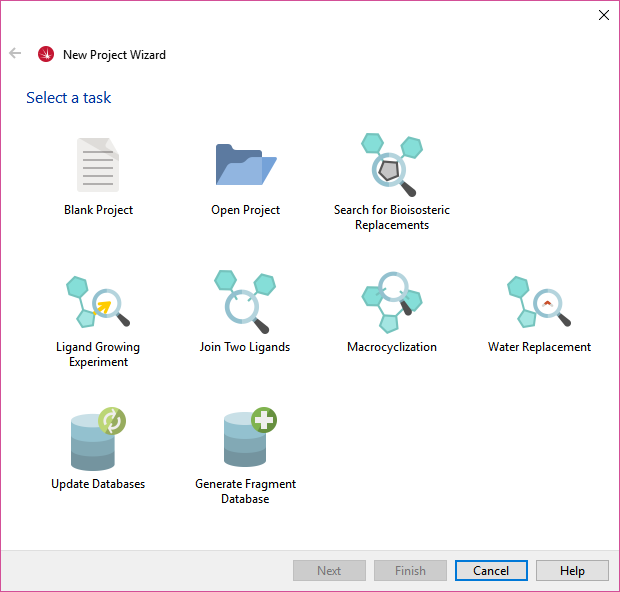
Figure 1. The new Spark project wizard.The ‘Ligand Growing Experiment’ wizard (Figure 2) can be used to grow a starter molecule into new space, guided by existing ligands mapping a different region of the same active site. This was possible in previous versions of Spark (see case study
Using Spark Reagent Databases to Find the Next Move) but the new wizard makes the workflow easier and more accessible.
The ‘Water Replacement’ wizard (Figure 2) can be used to search for a group which will displace a crystallographic water molecule near your ligand. Again the new wizard significantly improves the workflow for this popular Spark experiment that we have detailed previously (see case study Displacing crystallographic water molecules with Spark).
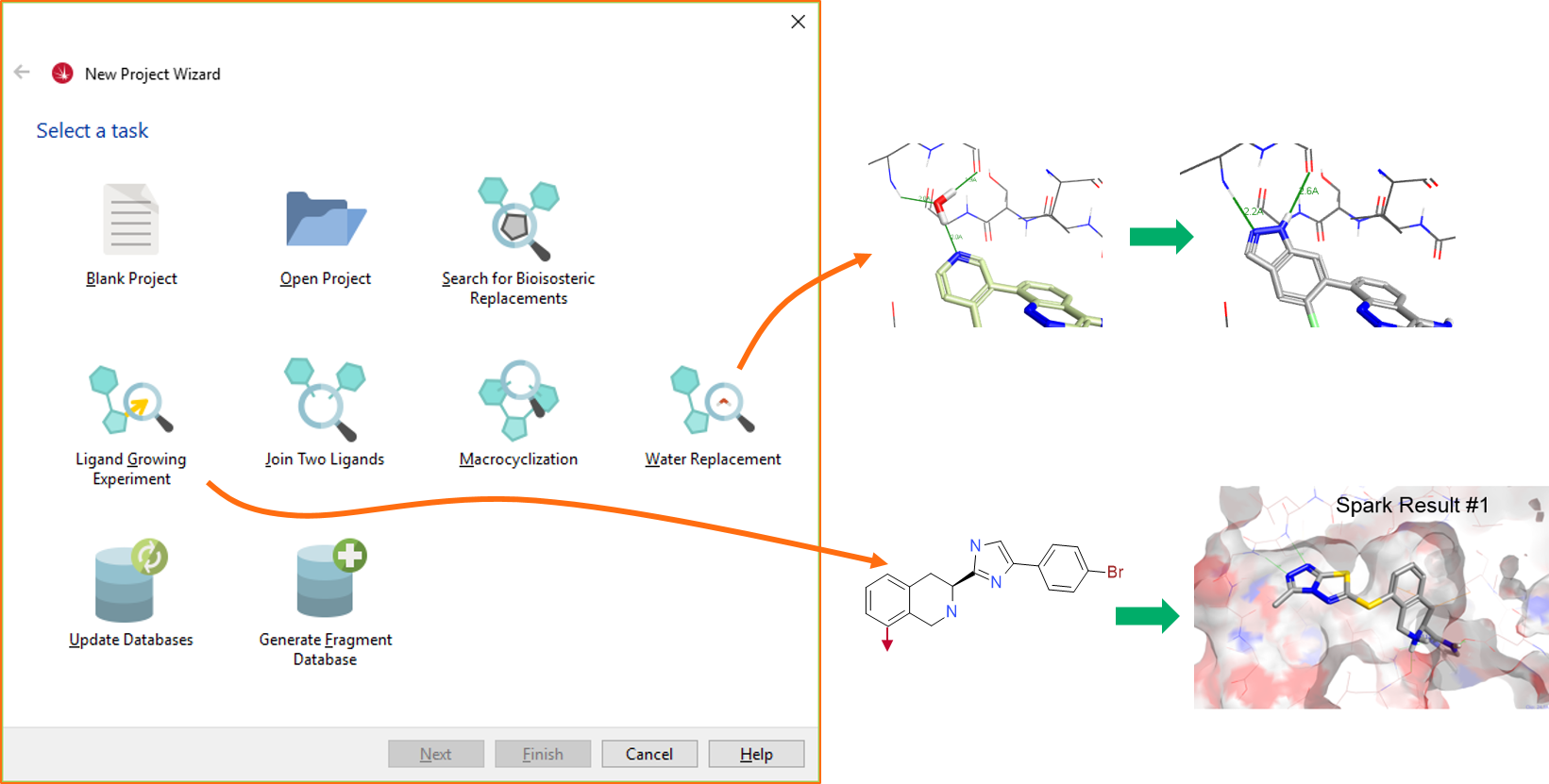
Figure 2. Results of ligand growing and water replacement Spark experiments.The ‘Join Two Ligands’ (to find a linker that joins two ligands sitting in the same active site) and the ‘Macrocyclization’ (to cyclize a molecule by joining two atoms with a linker) wizards are new workflows, which we have been testing internally in the last few years. Full case studies for these workflows are in preparation, but you can see an example of result you can get in Figure 3.
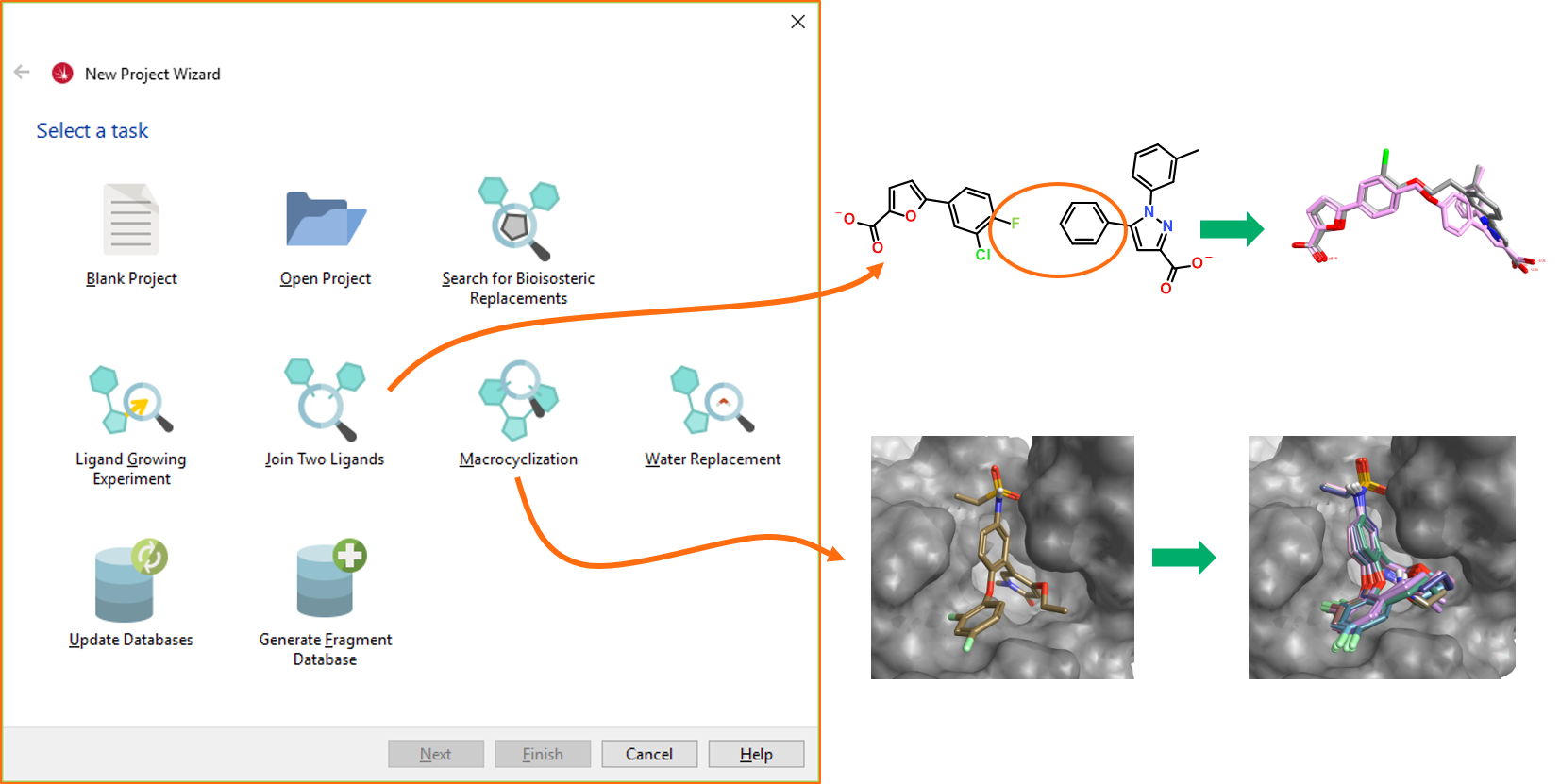
Figure 3. Results of joining two ligands and macrocyclization Spark experiments.In developing the wizards, a number of additional features to support these advanced experiments have been fine-tuned. These include:
- New ‘Ligand Growing’ and ‘Ligand Joining / Macrocyclization’ calculation methods to support ligand growing, ligand joining, macrocyclization and water replacement experiments
- A starter molecule sitting within a protein’s active site can now be downloaded directly from the RCSB
- Hydrogen atoms can now be replaced in the starter molecule
- New ‘merge’ functionality to merge molecules in the wizards (when appropriate) and in the Manage/Edit References dialog
- The starter molecule region selector now includes both 2D and 3D display.
These advanced bioisostere replacement experiments work well because of Spark’s product-centric approach. In Spark, result molecules are compared to the starter molecule, and a similarity score is calculated, only when the new molecules have been formed, minimized, and their fields and field points re-created. This approach, combined with the power of the Cresset XED force field, enables Spark to work with a higher level of precision, by avoiding fragment scoring limitations, allowing for neighboring group effects and for the electronic influence of replacing a moiety on the rest of the molecule and vice versa.
Enhanced Spark database update functionality
Spark V10.5 offers considerable improvements to the Spark database update functionality, to make this process more user friendly and efficient.
The Spark search dialog now alerts you when an updated version of the databases is available, before you start a search. Databases which need updating are marked by a green icon (Figure 4): and if the databases are selected for the Spark search, an ‘Update’ link appears at the bottom of the window, taking you directly to the Spark Database Updater. You will find this particularly beneficial if you use the eMolecules reagent databases where our monthly release schedule provides you with the latest availability information.
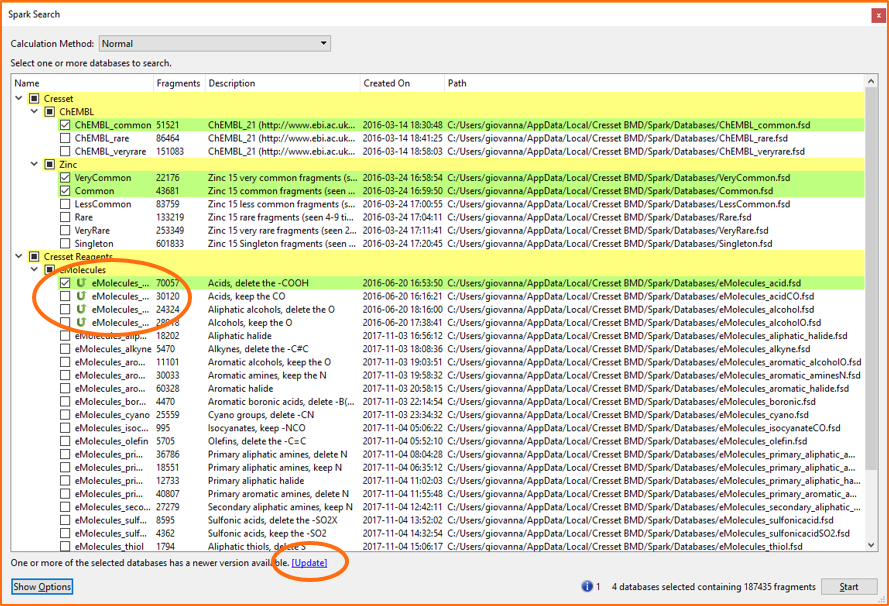
Figure 4. The Spark search dialog now alerts users when an updated version of the databases is available. If the databases which need updating are selected for the search, a link appears at the bottom of the window which takes you directly to the Spark Database Updater.The Spark Database Updater has also been improved. The databases are now categorized as ‘Cresset’ and ‘Cresset reagents’, to make it easy to locate those you wish to update (Figure 5). Furthermore, you can now download or update all the displayed databases in one go by clicking the ‘Install or Update Displayed Databases’ button at the bottom of the window.
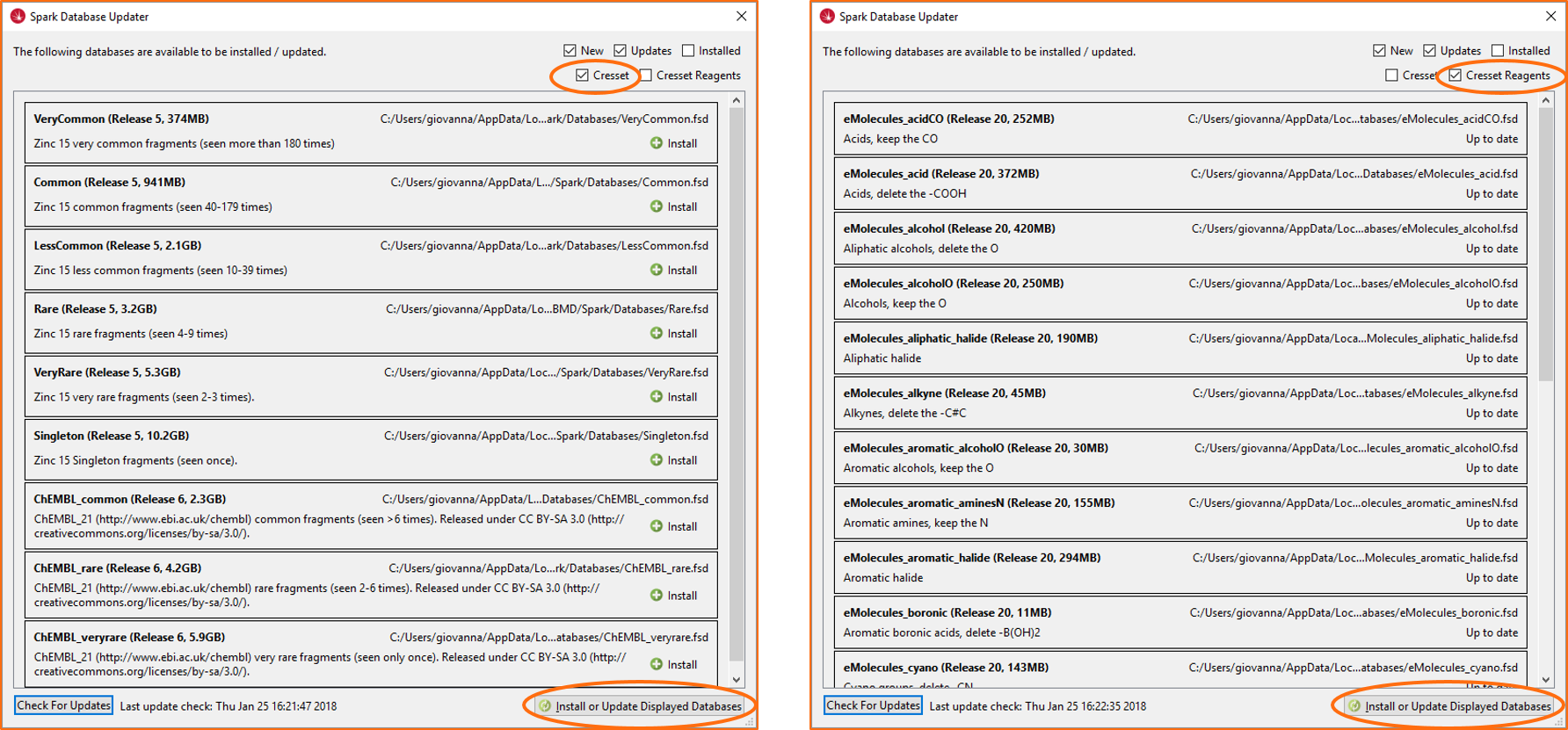
Figure 5. The Spark Database Updater now shows different categories of Spark databases and includes a button to install or update all the displayed databases in one go.Finally, a new ‘sparkdbupdate’ binary is available to enable you to update all or selected Spark databases from the command line.
What’s new in Spark searches
A significant number of improvements and new features to the Spark searches have been made in this release.
Field and pharmacophore constraints
Field and pharmacophore constraints can be used to bias the Spark search towards results which fit the known SAR or your expectations, by introducing a penalty which down-scores results that do not satisfy the constraint.
Field constraints enable you to specify that a particular type of field must be present in the Spark result. For example, you may want to a constrain a positive field where you want an interaction but this can be matched by both H-bond donors and other electropositive features such as the aromatic hydrogens of the compound in Figure 6 – right.
Pharmacophore constraints, new in Spark V10.5, ensure that the desired pharmacophore features are matched by an atom of a similar type in the Spark results. In Figure 6, a pharmacophore constraint was introduced to ensure that all Spark results contain a H-bond acceptor.
While field and pharmacophoric constraints are a powerful way of fine tuning Spark results, we recommend that they are using sparingly, as they will be introducing a bias in your experiment. For example, introducing a pharmacophore constraint on the indazole NH of the PDB 4Z3V ligand in Figure 6 – left would not have matched the aromatic hydrogens of the active ligand in Figure 6 – right.
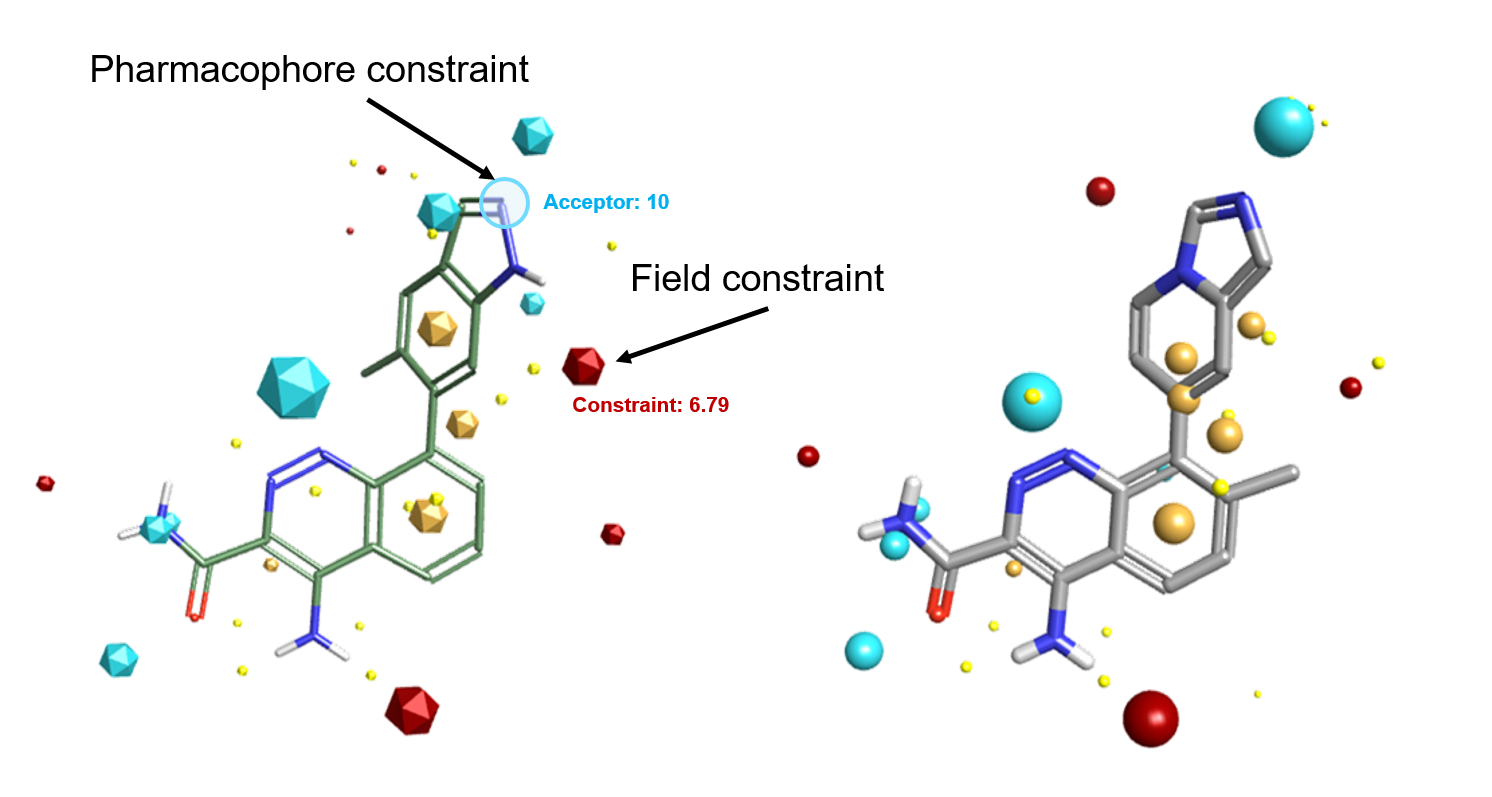
Figure 6. Left: Ligand from PDB: 4Z3V with pharmacophore and field constraints. Right: Active BTK ligand which satisfies both constraints.Read more about field and pharmacophore constraints in the
Forge V10.5 and
Blaze V10.3 release announcements.
Enhancements to the Spark search algorithm
Spark V10.5 also includes enhancements to the search algorithm and associated advanced options, for example:
- New functionality to weigh specific fields independently when scoring
- New similarity metrics to provide alternate scoring methods for the alignments
- New widget for adding field and pharmacophore constraints
- New ‘Flexibility’ filter which can be applied (together with SlogP and TPSA filters) to the whole molecule when performing a Spark search, to limit the results to the desired physico-chemical space.
Other new features and improvements
This Spark V10.5 release also includes a variety of additional new functionalities and improvements to the interface (Figure 7). These include:
- New ‘Send to Flare™’ functionality to send either all results, favorite results or selected results to Flare, including as appropriate the starter and reference molecule(s) and the protein
- New Storyboard window, to capture scenes recording all details from the 3D window that can be easily recalled when needed, including capability to annotate and rename scenes
- New stereo view functionality
- New support for touch screen displays and HiDPI
- New Flexibility column in Spark Results tables
- Improved performance of Spark database generation
- Improved ‘View Parent Structure for Selected Result’ functionality now including both substructure and identity search
- Improved ‘Grid’ button functionality
- Improved display of protein ribbons, offering a choice of different ribbon styles and capability to show ribbons for the active site only
- Improved look and feel of the GUI with re-designed toolbars and updated and clearer icons for a more modern and sleek interface.
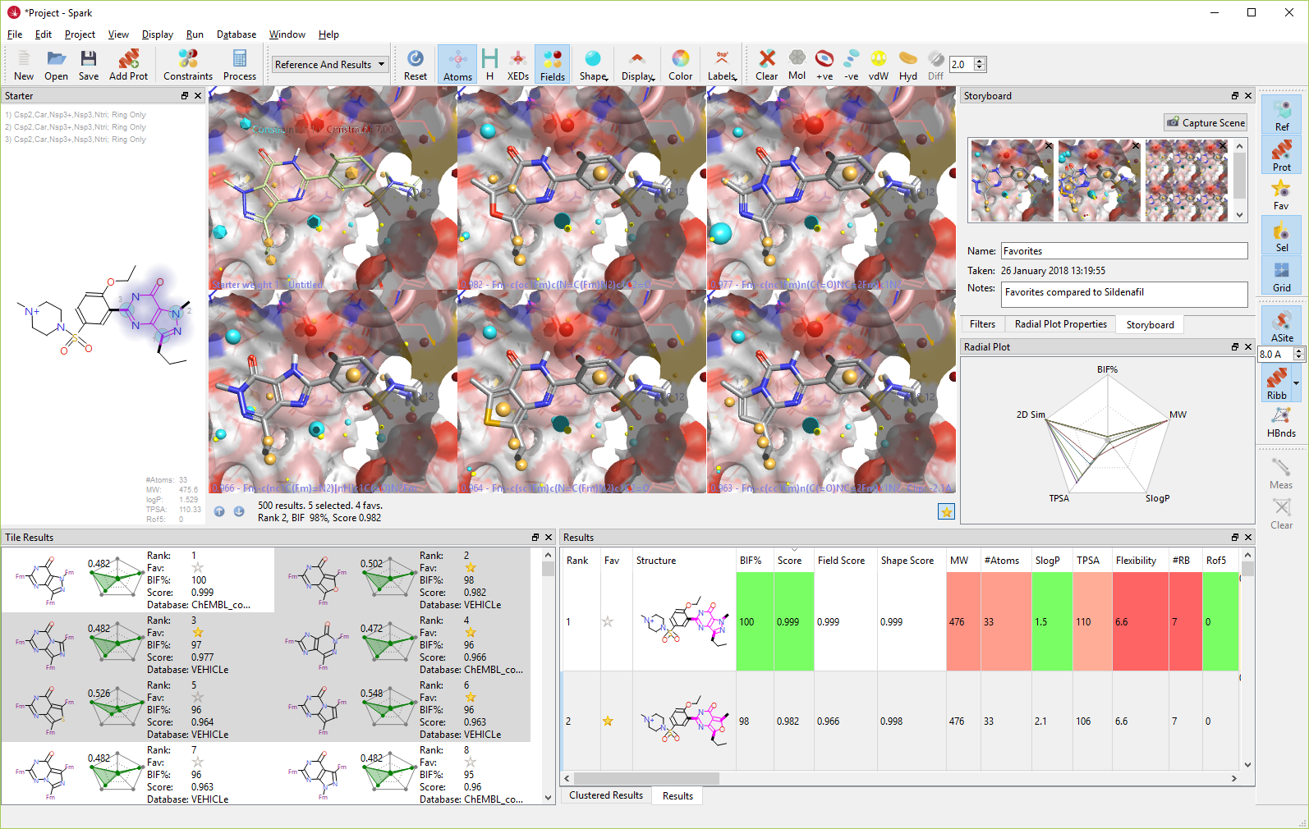
Figure 7. The Spark V10.5 GUI.
Try Spark V10.5
This release represents a significant improvement in the usability and flexibility of the leading bioisostere application. I encourage you to upgrade your version of Spark at your earliest convenience, and to download the keyboard shortcut guides for Spark V10.5 and Spark V10.5 Molecule Editor.
If you are not currently a Spark customer, please request a free evaluation.
Contact us if you have queries relating to this release.






