Future-proofing Cybersecurity in Drug Discovery
The pharmaceutical and biotech sectors suffer more data security breaches than any other industry, with 53% resulting from malicious activity. To protect against potential ...
News
We recently attended ICCS in Noordwijkerhout, where we presented two posters:
This was my second time at this conference, the first as a Cresset scientist. I remember very well my previous attendance in 2011, as it coincided with a turning point in my career: in fact, by that time I had realized that my main research interest had become the development and implementation of new methodologies in computational chemistry, and I started concentrating my efforts in that direction, ultimately joining Cresset a few years later.
Based on my two participations, I do rate ICCS very high in my personal ranking of scientific events, on par of the German and Sheffield conferences of cheminformatics: excellent scientific program, relaxed and informal atmosphere, extended poster sessions to merge and exchange ideas with colleagues, plus a sun-blessed boat trip on the Ijsselmeer.
In the following I will try to capture a few highlights of the talks that I found most inspiring, even though the list could indeed be much longer.
Greg Landrum (KNIME, T5 Informatics) presented a KNIME-based workflow for automated building of predictive QSAR models (Figure 1).
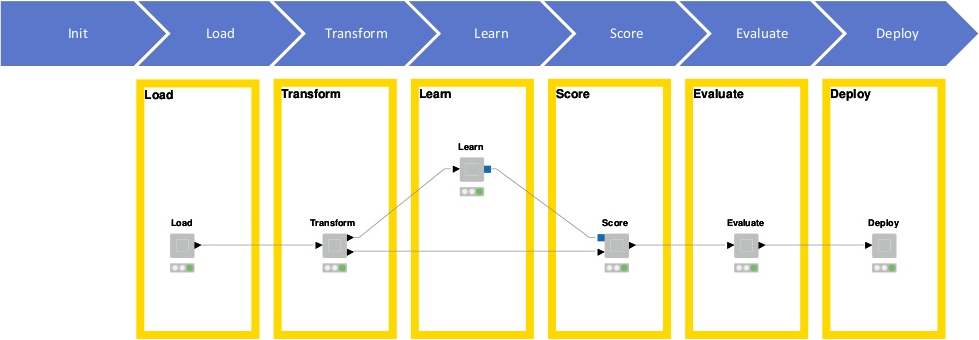
Figure 1. KNIME-based workflow for automated building of predictive QSAR models, presented by Greg Landrum, KNIME.
ChEMBL 23 was used as the data source for model training, after ‘seeding’ it with compounds roughly similar to the active compounds (i.e., Morgan2 fingerprint Tanimoto between 0.35 and 0.60) which are assumed to be inactive. In fact, the ratio of actives to inactives in scientific publications (from which ChEMBL data are extracted) tends to be unnaturally high, because researchers prefer to focus on active compounds in their reports. Five different kinds of chemical fingerprints were generated for each compound, and ten different activity-stratified training/test set splits were carried out for each activity type. Four different models were built using various machine learning (ML) methods, namely Random Forest, Naïve Bayes, Logistic Regression and Gradient Boosting; the best model was selected as the one giving the highest enrichment factor at 5%.
Overall, over 310K models were built and their performance evaluated in a fully automated fashion; calculations were distributed on 65-70 load-balanced AWS nodes and took only a couple of days. While results seemed initially deceptively good, more in-depth critical analysis revealed that most models were overfit on the training data and not very generalizable. This conclusion was very interesting in itself, adding up to the usefulness of the comparative analysis of the predictive performance of the various fingerprint/ML method combinations.
I enjoyed very much the talk given by Sereina Riniker (ETH) on deriving atom-centred partial charges for molecular mechanics calculations from ML models. Such models were trained on DFT electron densities computed with the COSMO-RS implicit solvent model (or COSMO for compounds containing elements not supported by COSMO-RS). Sereina had already anticipated that she was working on this subject during a lighting talk given at the 2017 RDKit UGM, and I had been looking forward to seeing the final results since then. The Random Forest model was trained on a set of 130K diverse lead-like compounds from ZINC/ChEMBL, picked as to include as many substructures as possible, with the goal to cover most of the known chemical space of drug-like molecules. In addition to being a lot faster than DFT, the methods yields 3D conformation-independent partial charges, which is a highly desirable feature (Figure 2).
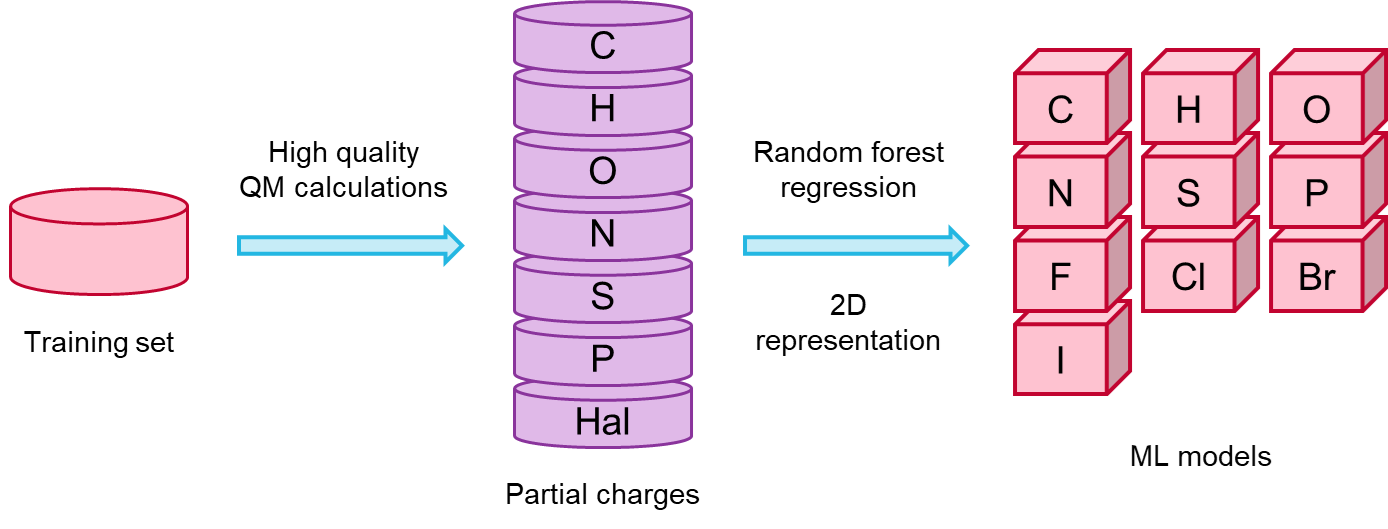
Figure 2. Procedure to derive 3D conformation-independent partial charges from ML models trained on QM data, presented by Sereina Riniker, ETH.
Prediction accuracy is >90%, and can be further improved by adding compounds to the training set in case some feature turns out to be poorly predicted due to being under-represented.
On a similar topic, Prakash Chandra Rathi (Astex) presented a method based on graph convolutional deep neural networks to predict the molecular electrostatic potential (ESP) around atoms in a molecule, with the goal of optimizing ligand-protein electrostatic complementarity, thus removing the need for expensive QM calculations. The neural network was trained on 48K diverse molecules for which ESP surfaces were computed using DFT calculations. The optimized model delivered excellent predictive performance on a validation set of 12K molecules (r2 0.95). In addition to visualization of field extrema, as in Cresset’s field point technology, ESPs computed in this fashion hold promise for developing knowledge-based scoring functions to estimate the propensity of a certain atom in a ligand to interact with another atom in a protein.
Knut Baumann (TU Braunschweig) described a method developed in collaboration with Bayer to deal with a very common problem in QSAR modeling, i.e., the fact that for weakly active compounds an exact pIC50 or pKi cannot usually be assessed, as it lies below the assay detection threshold. This kind of data is called left-censored, and poses a serious problem as regression algorithms able to handle hundredths or thousands of left-censored predictors are not available. Treating such data ‘as is’, i.e., interpreting the ‘lesser than’ symbol as if it were an ‘equals’ symbol or omitting this data altogether leads to underestimating the slope and/or overestimating the intercept. To address this issue, Baumann and co-workers adapted the Buckley-James algorithm to principal component regression (PCR) and partial least-squares regression (PLS). In a nutshell, a self-convergent procedure is adopted: the model is initially fitted against all data, then corrections to censored data are estimated, and after applying them the model is re-fitted and the whole procedure is iteratively repeated until convergence is reached. The method was validated using PLS and PCR on both simulated data and on a real dataset from industry, yielding a significant performance improvement compared to the naïve cases where left-censored data are handled as if they were uncensored and to the case where all left-censored data are simply neglected.
Bernd Kuhn (Roche) presented a thorough analysis of unusual intermolecular interactions (i.e., halogen bonding, dipole-dipole interactions of aromatic F, aromatic sulfur interactions, S σ-hole to carbonyl, etc.) as observed in crystallographic complexes from the Protein Data Bank and the Cambridge Structural Database. The goal of this study was assessing how important such interactions, on which a wide literature exists, actually are in real life. The analysis was done using the line-of-sight (LoS) paradigm (an interaction between two atoms can only take place if no other atoms are in the way), coupled with exposed surface areas to derive interaction statistics from crystallographic databases. 16 protein atom types were defined describing element, hybridization, polarity and charge of each atom, and their propensity to interact with other atom types was determined: for example, chlorine likes being an environment with polarized CH, C and NH π-systems, and non-polar groups, whereas cyano groups prefer hydrogen bond donors, positive charge, NH π-systems and waters. Additionally, the geometric preferences of each kind of intermolecular contact were determined.