Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
In a presentation at the Cresset User Group Meeting in 2016, Nik Stiefl and Finton Sirockin from Novartis discussed the selection of building blocks for DNA encoded libraries using electrostatic and shape diversity as the key descriptor. This work was powered by a custom binary and scripts written by Cresset. Over the last couple of years this approach has been applied to a wider range of library designs and has gained a reputation as a method of choice for many library designs.
 PickR will be a new tool that formalizes the approaches that we developed in collaboration with Novartis. It is a command line binary that provides a diverse pick of reagents to be incorporated into a library. Unlike other approaches, PickR uses the electrostatic and shape properties of molecules to generate the descriptor matrix that is used as the basis for the diversity pick.
PickR will be a new tool that formalizes the approaches that we developed in collaboration with Novartis. It is a command line binary that provides a diverse pick of reagents to be incorporated into a library. Unlike other approaches, PickR uses the electrostatic and shape properties of molecules to generate the descriptor matrix that is used as the basis for the diversity pick.
Generating diversity using a 3D property is not straight forward. It is necessary to explore conformations of R-groups and rotate them about the proposed connection point in order to fully understand the distribution of properties. Much of this was well described in Nik and Finton’s presentation (slides 12-15). I will leave a formal discussion to the final release announcement.
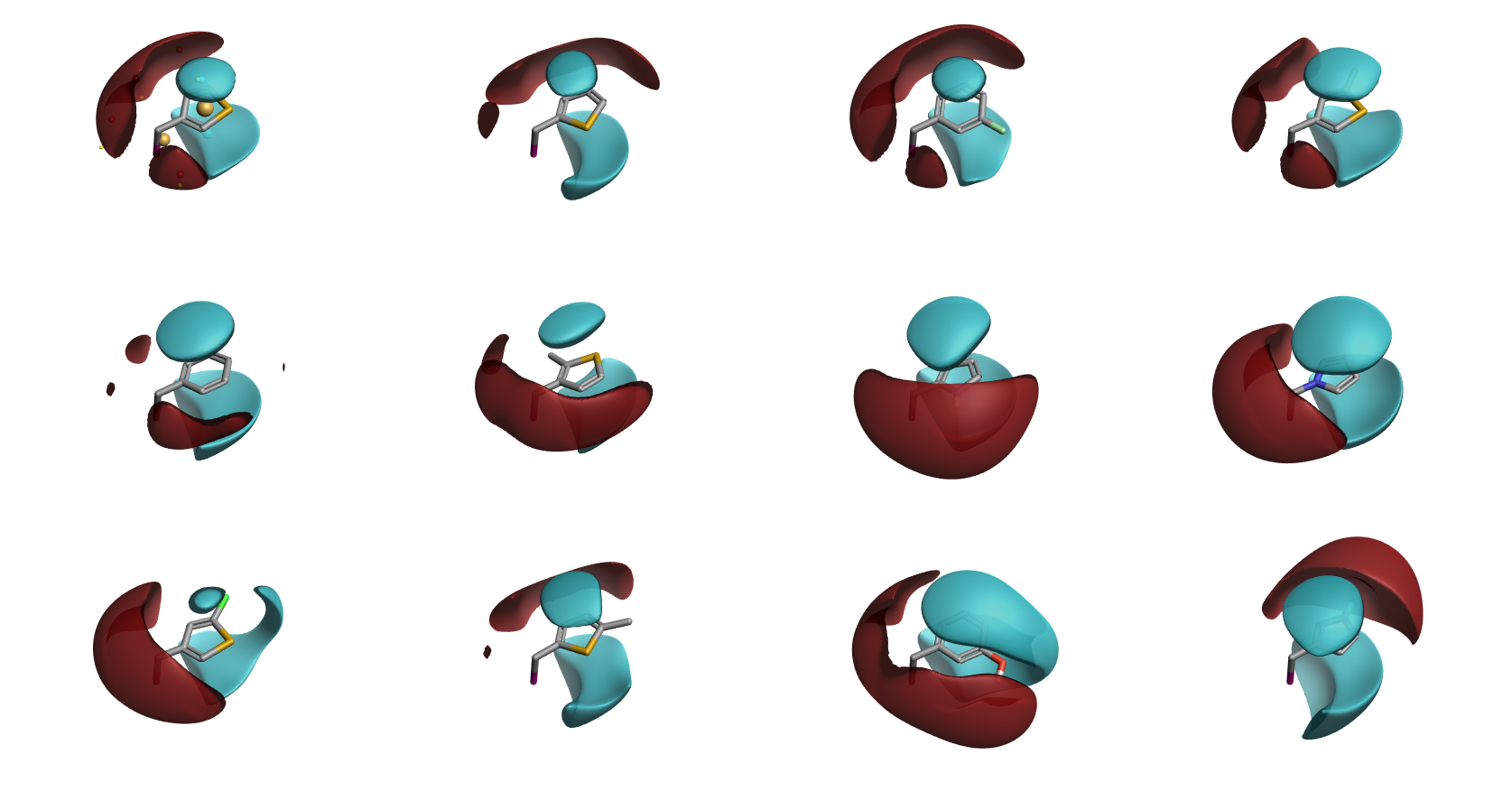
I applied a dataset of approximately 1,000 amino acids that can be purchased from eMolecules. The raw reagents were processed to convert the side chains into R-groups with the C-alpha atom being converted to Iodine (all other Iodine containing reagents were excluded as were those containing Br and those with side chains >150Da). Using PickR, I generated a 3D similarity matrix for the side chains, aligning on the C-alpha to C-beta bond. 100 clusters were requested initially.

3D representations of all 100 clusters generated from amino acid side chains, aligned to each other using the I-C bond of the fragments.
Looking at the results, there are some very nice relationships. For example, in Cluster 2, together with tyrosine, are other phenolic side chains but also an indazole that contains the donor-acceptor motif.
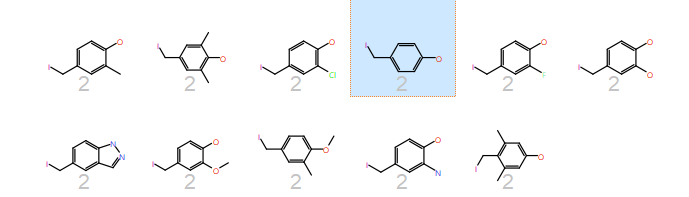
2D representations of all the side chains in the same cluster as tyrosine (highlighted).
Along with the indole of trytophan are other substituted indoles, pyropyridines and benzofuran. In with the isobutyl side chain of leucine are a number of cyclic analogues which I expect would cause issues with many 2D similarity methods. Interestingly, indoline is placed together with the equivalent of homo-phenylalanine.
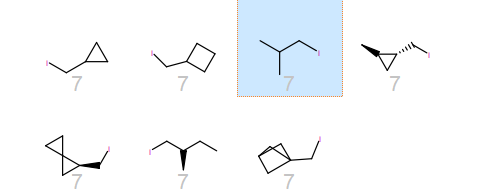
2D representations of the leucine related cluster.
2D representations of the homo-phenylalanine related side chains
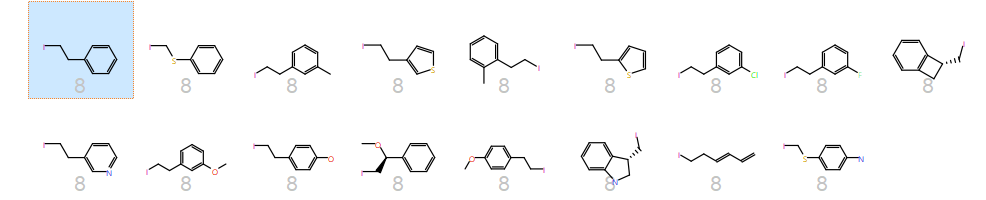
The cluster containing the phenylalanine side chain highlights the major difference of PickR over other methods – R-groups are clustered on 3D electrostatic properties. Hence, together with the phenylalanine side chain you have thiophenes and pyroles but few other aromatics – pyridine and pyrimidines go to their own clusters because using electrostatics they are quite different to a plain phenyl ring.
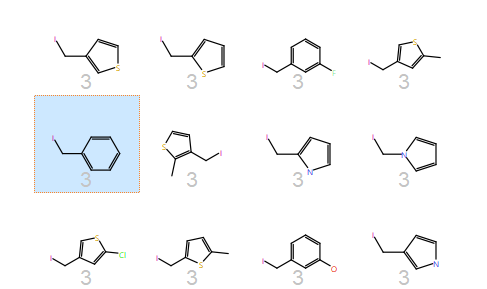
3D and 2D pictures of all side chains in the phenylalanine cluster
Contact me to receive the full results in a Forge project file.
Contact your account manager if you are interested in learning more about PickR.