CCPBioSim (Collaborative Computational Project for Biomolecular Simulation) is an inclusive and wide ranging project, bringing together chemists, physicists and chemical engineers as well as researchers from all branches of ‘molecule-oriented’ biochemistry and biology.
Earlier this month I attended the CCPBioSim training week, hosted by the University of Bristol. The workshops were aimed at training biomolecular researchers in current methodologies and tools in biomolecular simulation. Training material was mainly presented in the form of interactive Python Jupyter notebooks, neatly combining illustrated instructions, code examples and exercises, and data visualization (Figure 1).
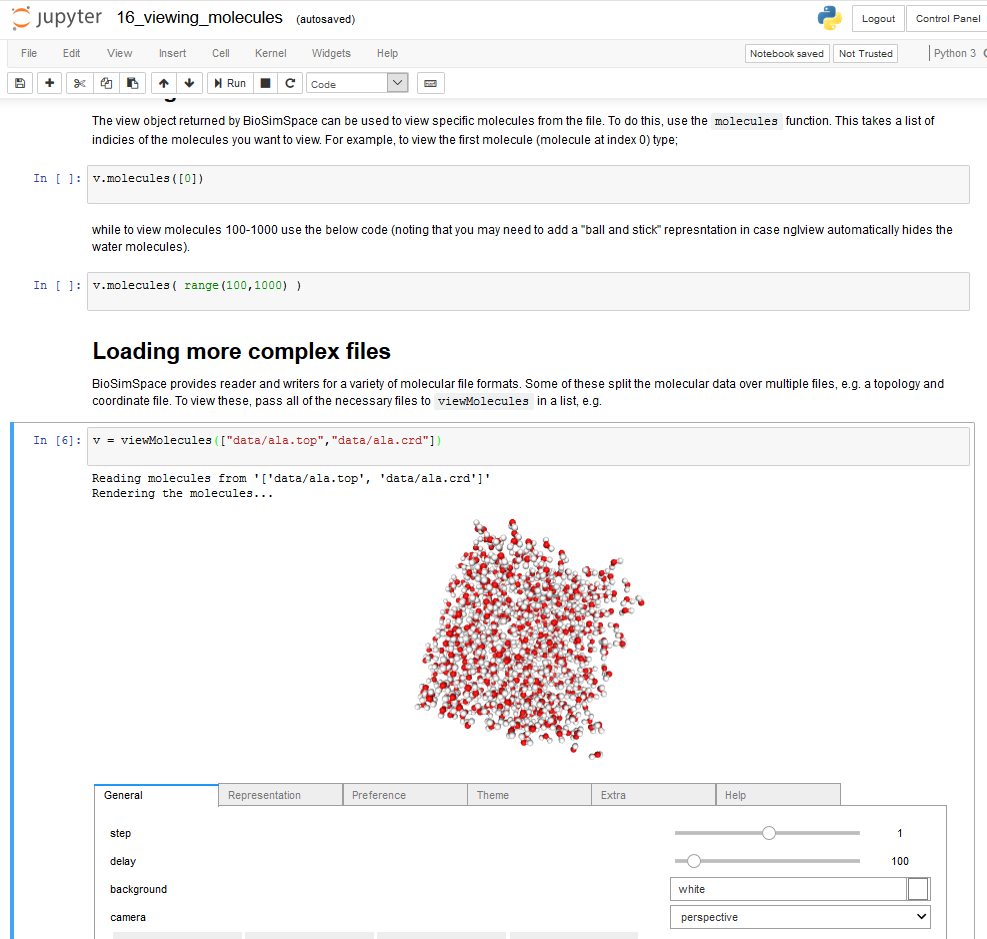
Figure 1: Writing python code and visualizing molecules in an interactive Jupyter notebook.
The workshops covered:
- Introduction to Python for biomodelers, by Dr Christopher Woods (University of Bristol) who taught the basic skills and knowledge to write python code in the context of biomolecular simulations
- Good practice and pitfalls in setting up, running, and analysing molecular dynamics simulations, by Professor Charles Laughton (University of Nottingham). This session featured a practical example of selecting suitable NMR structures using the PROCHECK webserver and an introduction to MD trajectory analysis methods using the python modules MDtraj, matlabplotlib, and pyPcazip
- Introduction to setting up alchemical free energy perturbation calculations with FEsetup1 and Sire SOMD, by Dr Antonia Mey (University of Edinburgh). Hydration free energies of ethane and methanol and relative ligand binding free energies to lysoszyme were set up and calculated
- Overview of different methods and best practices for analysing alchemical free energy calculations and computing protein-ligand binding free energies in an interactive python environment using ‘analyse_freenrg’ function of Sire
- ISAMBARD2, an open-source Python package for structural analysis and rational design of biomolecules, was used to analyze structural elements of proteins and de novo-model coiled-coil structures
- BioSimSpace, a large collaborative open-source project comprising multiple academic groups and industrial partners, including Cresset. BioSimSpace aims to remove barriers of different input formats and data preparation between different molecular simulation packages, enabling workflows to seamlessly plug together. Furthermore, it is usable in diverse interactive environments such as Jupyter notebooks and KNIME and provides a community hub and repository for developing software and biomolecular workflows together.
- Examples of running free energy calculations with WaterSwap3,4 (integrated in Cresset’s Flare5), LigandSwap, and ProteinSwap (all part of Sire) for predicting how changes in a ligand or mutations of a protein affect protein-ligand binding. These methods also provide an in-built residue-based decomposition of the binding free energy, which makes it possible to visualise and rationalise predicted changes in binding affinity according to changes in specific protein-ligand interactions (Figure 2).
- Introduction to QM/MM modeling of enzyme reactions, by Dr Marc van der Kamp (University of Bristol). During this workshop theoretical background, best practices for set up and analysis, and choice of QM level were discussed. As practical example, the AmberTools package was used to calculate the potential and free energy surface of the chorismate mutase catalysed reaction of chorismate to prephenate using the semi-empirical AM1 QM method.
- Introduction to the ProtoMS software package and running biomolecular Monte Carlo simulations, by Dr Christopher Cave-Ayland and Hannah Bruce-Macdonald (University of Southampton). Subsequently, a very interesting application of ProtoMS was presented and run: The calculation of water affinities in protein binding sites with Grand Canonical Monte Carlo simulations.6,7 These simulations run at a µVT ensemble (constant chemical potential) and include insertion and deletion moves of water molecules, allowing to perform a titration of the chemical potential to determine free energies of each water molecule in the respective binding site.
All workshop attendees were given the opportunity to participate in an exciting virtual reality docking experiment, allowing to actually move ‘inside’ a biomolecular simulation while docking a small molecule ligand into a protein pocket with VR hand controllers.
Overall, the well-organized CCPBioSim training workshop laid a great foundation for good practices with set up, running, and analysis of biomolecular simulations and showcased exciting developments and applications. The whole week had a very collaborative and supportive atmosphere. See for the Twitter discussion.
Thank you to the organizers, speakers, and attendees that have made this workshop so enjoyable and a great success.
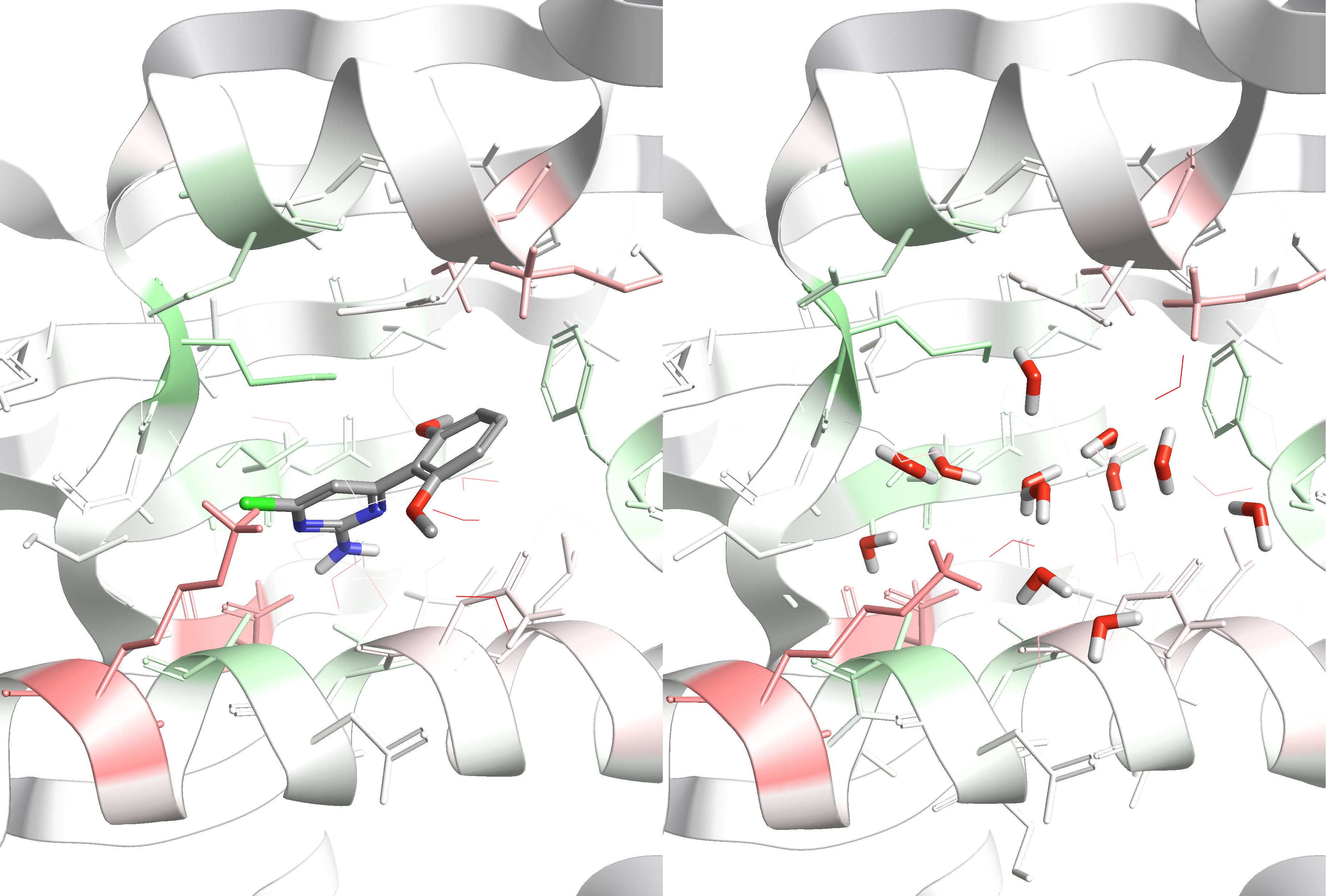
Figure 2: Visualization of residue-based decomposition of the protein-ligand binding free energy of WaterSwap calculations in Flare5 (HSP90 example, pdb code 2XDK). Green areas highlight residues that prefer interactions with the ligand (left panel), whereas red areas highlight residues that prefer interactions with the water cluster (right panel).
References
- Loeffler, H. H.; Michel, J.; Woods, C. FESetup: Automating Setup for Alchemical Free Energy Simulations. J. Chem. Inf. Model. 2015, 55 (12), 2485–2490
- Wood, C. W.; Heal, J. W.; Thomson, A. R.; Woolfson, D. N. ISAMBARD: An Open-Source Computational Environment for Biomolecular Analysis, Modelling and Design. Bioinformatics 2017, 33 (19), Pages 3043–3050
- Woods, C. J.; Malaisree, M.; Hannongbua, S.; Mulholland, A. J. A Water-Swap Reaction Coordinate for the Calculation of Absolute Protein–ligand Binding Free Energies. J. Chem. Phys. 2011, 134 (5), 054114
- Woods, C. J.; Malaisree, M.; Michel, J.; Long, B.; McIntosh-Smith, S.; Mulholland, A. J. Rapid Decomposition and Visualisation of Protein–ligand Binding Free Energies by Residue and by Water. Faraday Discuss. 2014, 169, 477–499
- Flare
- Cave-Ayland, C.; Skylaris, C.-K.; Essex, J. W. A Monte Carlo Resampling Approach for the Calculation of Hybrid Classical and Quantum Free Energies. J. Chem. Theory Comput. 2017, 13 (2), 415–424
- Ross, G. A.; Bruce Macdonald, H. E.; Cave-Ayland, C.; Essex, J. W. Replica-Exchange and Standard State Binding Free Energies with Grand Canonical Monte Carlo. J Chem Theory Comput 2017, 13 (12), 6373–6381.

