Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Here at Cresset we’re very interested in ligand-based virtual screening – it’s been a focus of the company ever since we started more than seventeen years ago. In that time there have been many advances and refinements of the techniques for both ligand-based virtual screening and structure-based methods. We have stuck by our fundamental principle that ligand similarity based on both electrostatics and shape is an excellent way to sort the wheat from the chaff. The results obtained by our services division, who have run more than 200 virtual screening campaigns with a better than 80% success rate, is testament to that.
One of the things our customers ask from time to time is which application should they be using to do virtual screening. The simple answer is that there are two, Forge (and its command-line utility ‘falign’) and Blaze, and the differences are readily apparent.
In falign, you can generate conformations for a large set of molecules, align them to one or more references, and rank them by the similarity score. You also have the option to bias the alignments and scores by adding field constraints, pharmacophore constraints, and protein excluded volumes.
By way of contrast, in Blaze, you can generate conformations for a large set of molecules, align them to one or more references, rank them by the similarity score, and… ok, point taken. So, given that falign and Blaze apparently do the same thing…
The answer is scale. As anyone who’s ever played with large data sets knows, doing calculations on a few hundred compounds is fundamentally different to doing them on tens of millions of compounds. Once you are working at large scale, seemingly trivial operations such as filtering data sets become much more difficult if you want to be efficient. Blaze was designed from the ground up to work with large data sets of 107 molecules and more, with an emphasis on maximizing throughput on a computational cluster. Forge/falign on the other hand are much more aimed at small-scale work, enabling simple screening or analysis of relatively small sets of compounds where the big iron of Blaze is overkill.
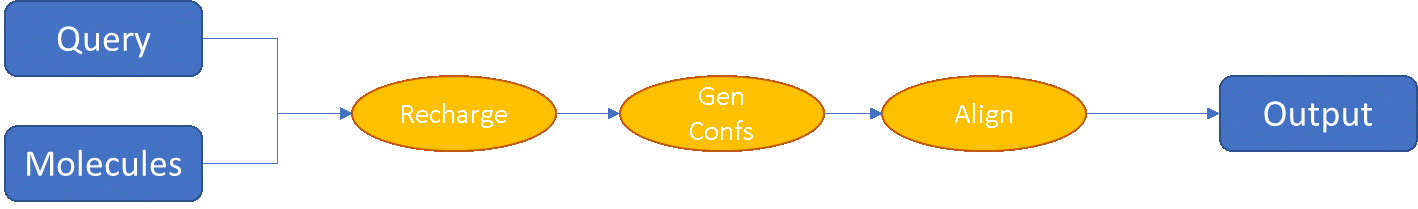
As an example, let’s look at the preparation of the data set in the two software suites. In falign, this is relatively simple: you provide the compounds to falign in 2D or 3D form, it assigns protonation states as necessary, and computes conformations on-the-fly if required before aligning to the query:
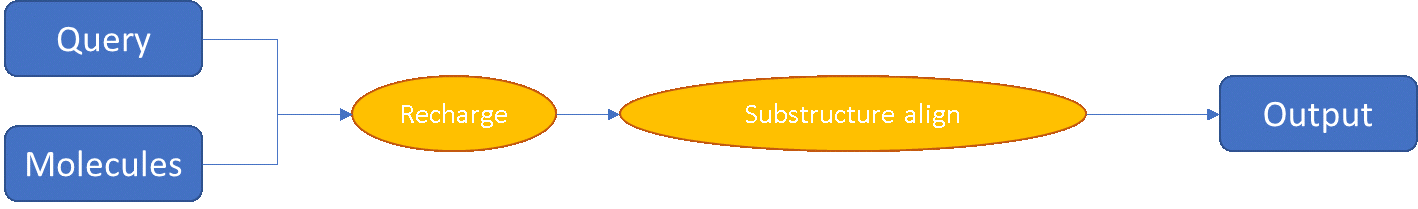
Falign has a secondary mode for use when aligning structurally-related compounds, which ensures that the common substructure within the dataset is perfectly matched:
Blaze, on the other hand, is much more sophisticated in its conformer handling. The average user of Blaze has multiple data sets that they want to screen (in-house compounds, vendor screening compounds, virtual libraries, custom collections), and these often have significant overlap. In addition, these data sets are usually reused multiple times for multiple virtual screens. As a result, Blaze has a sophisticated deduplication and precomputation pipeline that maximizes computational efficiency. The Blaze workflow looks more like this:
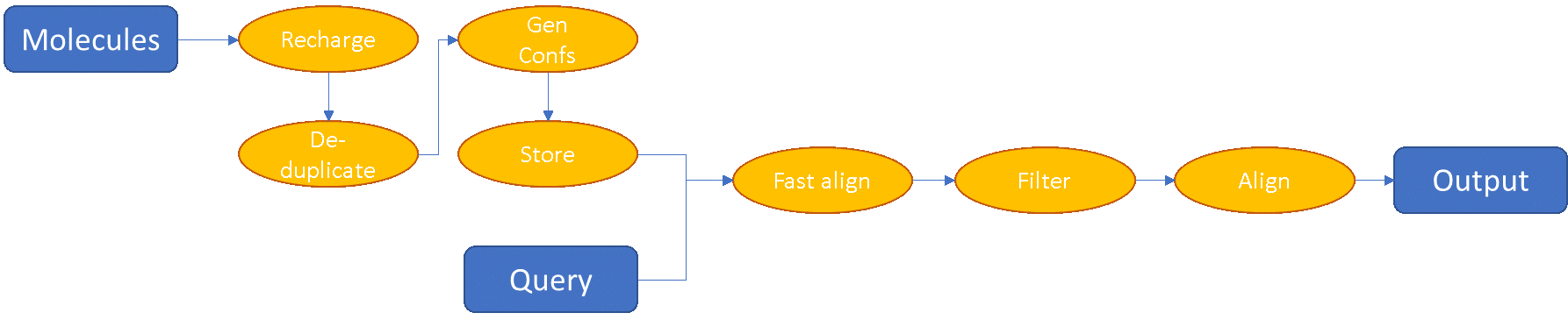
Any given chemical structure is only present once within Blaze: it may have multiple different names, and be present in multiple collections, but we’ll only precompute its conformations once and we’ll only align it once in any given screen. The conformer computation pipeline is heavily optimized for performance: we’ve done extensive studies on our conformer generation algorithm XedeX to find the optimal trade-off between conformation space coverage, rejection of higher-energy conformations, calculation time and number of conformations required. In addition, we’ve developed a special-purpose file format that is highly compressed (less than 13 bytes per atom on average, including coordinates, atom types, element, charge and formal charge) while being unbelievably fast to parse.
Blaze has a multiple-step pipeline to filter the data set, so that the full 3D electrostatic shape and alignment algorithm is only applied to molecules that are likely to have a high score. For extremely large data sets there’s an initial filter by FieldPrint, an alignment-free fingerprint method that gives a crude measure of electrostatic similarity. The molecules that pass the filter then go into an ultrafast version of our 3D alignment and similarity algorithm, and the full similarity algorithm is applied only to the best 10% or so of these. As a result, Blaze can chew through millions of molecules very quickly on even a modest cluster. The processing capability of Blaze is further enhanced by the fact that there’s a GPU version which is even faster.
So, falign is designed for the simple use case on small sets of molecules, while Blaze is aimed at maximum computational and I/O efficiency on very large data sets. There is another important difference between the two. As anyone who’s been in charge of maintaining a virtual screening system knows, keeping it up to date is often a painful and thankless task. It’s bad enough keeping up with the weekly additions to the internal compound collection but keeping track of updates to external vendor’s collections is difficult: not only are new compounds being added but old ones are being retired. Blaze makes handling this situation easy. You simply provide Blaze with the new set of compounds that you want to be in the collection, and Blaze will automatically handle the update.
Any new compounds will be added, no-longer-available compounds will be marked and removed from the screening process, and any unchanged compounds will be left alone. This is far more computationally efficient than fully rebuilding the conformations for everything. Blaze can even be directly connected to your internal compound database, so that the Blaze collection holding your in-house compounds is always right up to date.
Blaze is optimized for throughput and computational efficiency, but the downside of this is latency. If you have a set of compounds you want to align and score in Blaze, you have to upload them, wait for Blaze to process them and build the conformations, wait for Blaze to build its indices, initiate a search, and wait for it to be submitted to your cluster queueing system. There’s five- or ten-minute’s latency in all of this, which is fine for a million molecules but is overkill if you have only one hundred. Falign, by contrast, will start work straight away on your local machine with no waiting at all.
The answer to the falign vs Blaze question, then, is largely a question of scale. Got a dataset of a million molecules that you want to run repeated virtual screens against? Blaze is just the ticket. Got a small set of compounds that you want to align and score as a one-off? Forge and falign are just what you need. For our in-house work we tend to find that the tipping point occurs around a few thousand molecules. Falign can easily chew through this many in an hour or so (especially if plugged into your computing cluster using the Cresset Engine Broker). However, if there’s more compounds than this or we’re going to want to run multiple queries, then Blaze it is. Since Blaze is accessible through the Forge front end, and both are accessible through KNIME and Pipeline Pilot, it’s as easy as pie to pick the right tool for the job.
Request a free evaluation to try out Forge or Blaze for your small or large-scale virtual screening needs. Don’t have a cluster? Blaze Cloud and Blaze AWS provide simple ways to access cloud resources to do the number crunching for you.