Antibodies (immunoglobulins) are a key component of the mammalian immune response and are increasingly of interest as a means of therapy for several diseases. In fact, there are over 100 monoclonal antibodies that have been approved as drugs1. Despite this apparent success, there are issues with biopharmaceutical antibody research and development – not least the cost of goods. Although it can be argued that therapeutic antibody development relies on semi-random search-and-selection methods, such as phage display and biopanning, there is an increasingly important role played by structure-based design. For instance, the recent development of bispecific antibodies, where two antigen binding domains are incorporated into one antibody, requires careful design of the linker.
In late 2022 there are 2894 PDB entries listed in AbDb, a database of antibody structures2 and 6685 antibody structures listed in SAbDab3. This huge corpus of data on one protein family provides a trove of information on how antibodies interact with their antigen targets at the atomic level. Our knowledge of antibody structure-function relationships has allowed the engineering of antibodies to improve characteristics such as antigen affinity and effector function. The evolution of computational tools for protein modeling has also improved our ability to model antibodies.
Cresset Discovery has extensive experience in protein and antibody modeling. Our team of CADD experts can provide a deep insight into your antibody system from a molecular perspective by applying specialist computational approaches and expertise.
A case for antibody engineering
B cells produce antibodies in response to the presence of antigens. These antibodies then recognize and bind to antigens, triggering one of several downstream activities, such as:
- Neutralization by alteration of the chemical composition of an antigen
- Immobilization of the antigen-presenting entity
- Stimulation of the complement system leading to lysis of antigen-presenting cells
Memory B cells and antibodies for a specific antigen can then reside within the body for prolonged periods after exposure to the antigen as a mainstay of host defense.
Engineering antibodies can serve as a method of improving the therapeutic behavior of antibodies beyond what their wildtypes are capable of. For example, an antibody can be engineered to improve binding affinity to a particular antigen, to conjugate a drug, or to prolong the time antibodies reside in circulation. Each of these use cases requires identification of the antibody component that directs that particular action. In the case of improving antigen affinity, it is necessary to know and understand the residues present on the variable light and heavy chains and the interactions made between the antibody and the antigen.
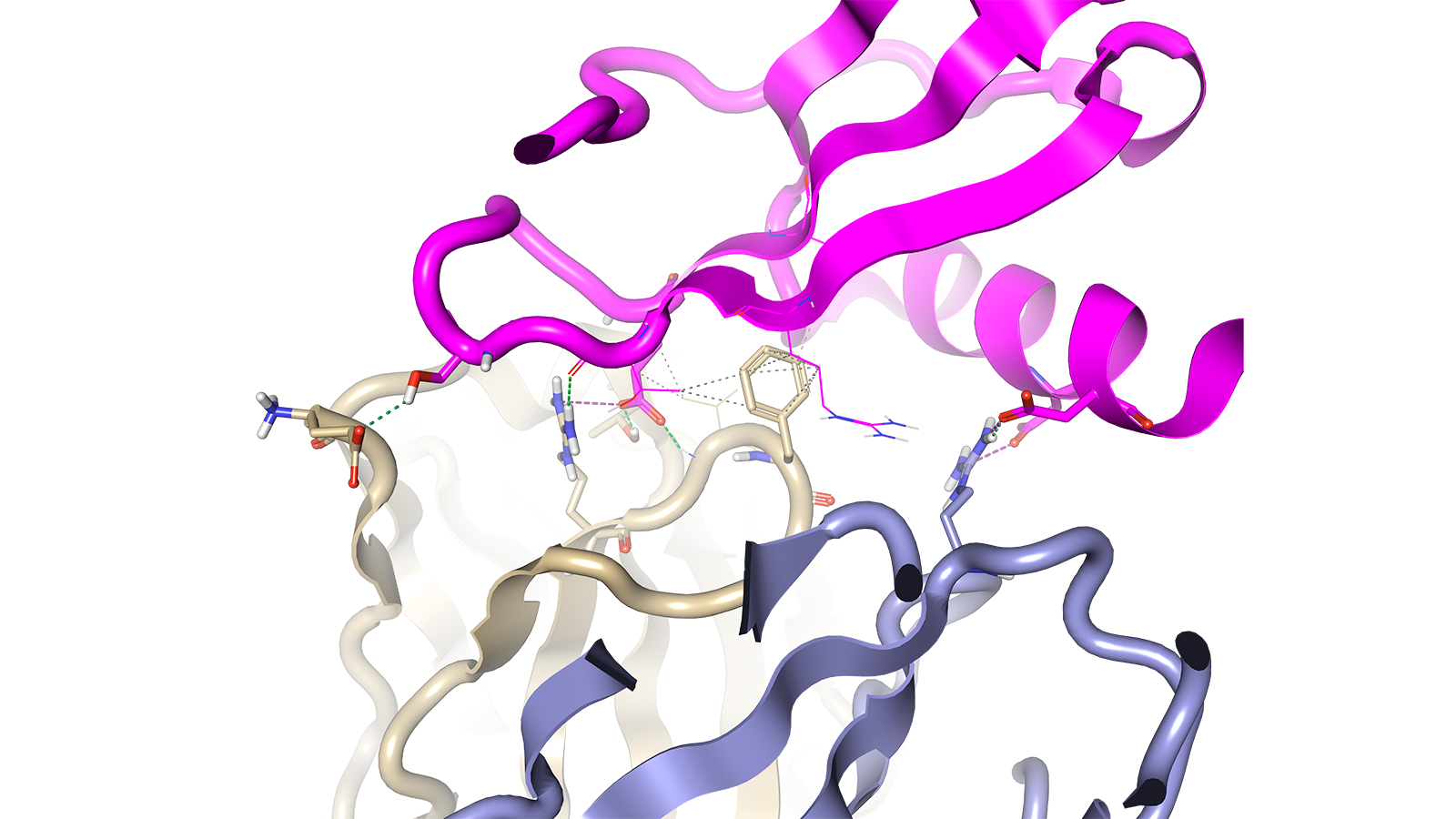
Figure 1. Select interactions between the heavy (beige) and light (blue-grey) chains of wild type antibody scFv 11K2 and the antigen MCP-1 (magenta) PDB ID: 2BDN.
Computational approaches to antibody engineering
Cresset Discovery applies sophisticated computational tools to visualize and understand antibody-antigen interactions, such as which residues contribute to the formation of the complexes. Once the antibody region of interest has been identified, our team can visualize the proximities of certain residues with others. Residues in the wild type antibody that are in close proximity with the antigen but aren’t eliciting favorable interactions with the antigen can serve as potential targets for a mutagenesis study.
Molecular representation allows visualization of interacting residues within the complex and can rationalize decisions about which residues to mutate. Residue mutations can also be performed quickly and effectively by Cresset Discovery.
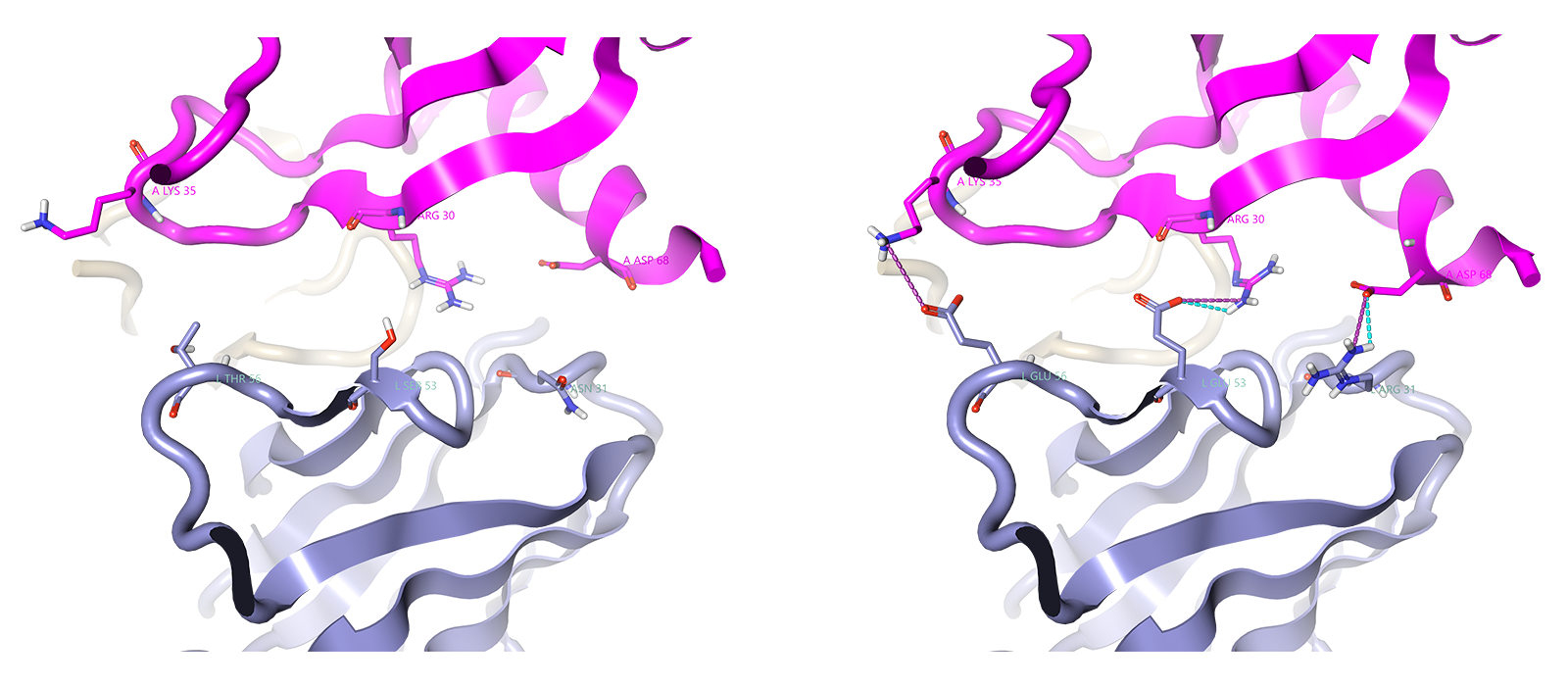
Figure 2. Example residue mutations performed computationally at the antibody light chain. Mutations from left to right: T56E, S53E and N31R. a) shows the wildtype residues in close proximity to polar antigen residues, b) shows the mutations at those antibody residues after a few rounds of energy minimization.
Figure 2 shows a Flare representation of residues at the antibody interaction site that are close to those at the antigen surface but are incapable of engaging in electrostatic interactions. These have been mutated into residues that could forge new interactions. Kiyoshi et al.4 found that mutating these three residues established three new electrostatic bonding interactions between the scFv 11K2 antibody and antigen MCP-1, and therefore improved affinity. The authors identified three amino acids – Thr56, Ser53 and Asn31 – that were not part of the standard antigen-antibody interface, but seemed to be destabilizing the interaction as they were not complementary with the antigen local surface. The electrostatic bonding interactions after mutating Thr56 to Glu, Ser53 to Glu and Asn31 to Arg can be seen in Figure 2b after a few rounds of energy minimization.
Computational analysis of antibody-antigen interactions
As well as mutagenesis and other protein engineering experiments, computational approaches give insight into the effects of these changes. One way of doing this is by investigating the effect that these new changes have on the interaction between the two proteins. Changes to residues on the antibody surface will also change the surface electrostatics, something that Cresset Discovery scientists can analyze with ease using Flare™, Cresset’s proprietary molecular modeling platform. Surface electrostatics show whether or not the antibody residue changes are more or less complementary to nearby antigen residues.
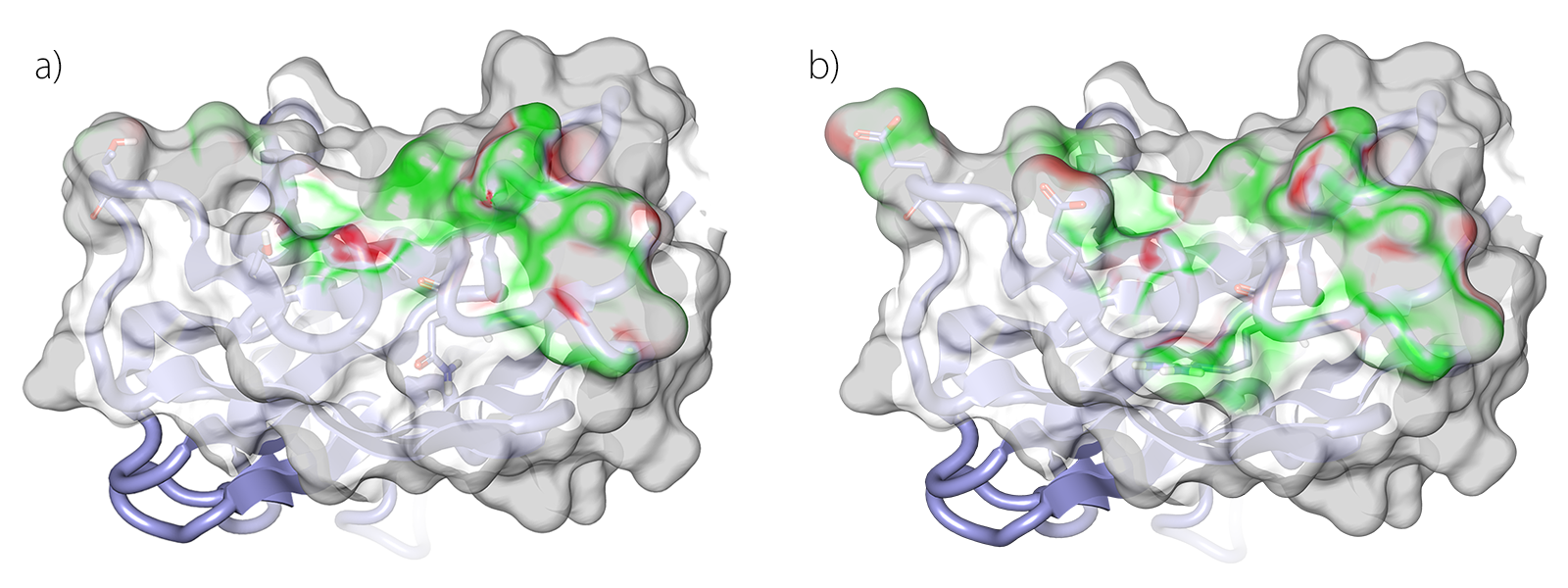
Figure 3. Electrostatic Complementarity (EC) surfaces of the wildtype (a) and mutant (b) antibody scFv 11K2 with respect to the antigen MCP-1.
Figure 3 shows an example of how antibody-antigen complex interactions can be analyzed using Electrostatic Complementarity™. Surface areas of close contact between antibody and antigen are color-coded green for attractive, i.e. complementary electrostatics. Red areas show where the antigen and antibody surfaces have a similar charge and so have a repulsive interaction. Figure 3(a) shows the original antibody with some complementary surfaces, but 3(b) – the mutated form – has a more extensive area of green, because of improved electrostatic complementarity between mutant residue and antigen. Our Cresset Discovery scientists use state of the art protein modeling in the Cresset suite of software to easily assess and predict the result of a mutagenesis investigation.
Engage Cresset Discovery to accelerate your project
Cresset Discovery serves as a true partner for computational modeling in drug discovery. Our expert team has a breadth of experience across medicinal and computational chemistry, and biology to work alongside you to deliver your project goals.
To learn more about how we can support you in an antibody engineering project or any other project, contact us or request a no-risk free confidential discussion with our team.
References
- Manis, J. P. “Overview of therapeutic monoclonal antibodies”, https://www.uptodate.com/contents/overview-of-therapeutic-monoclonal-antibodies.
- Ferdous, S.; Martin, A. C. R. “AbDb: Antibody Structure Database - A Database of PDB-Derived Antibody Structures”, Database 2018, Article ID bay040, 1. https://doi.org/10.1093/database/bay040.
- Dunbar, J.; Krawczyk, K.; Leem, J.; Baker, T.; Fuchs, A.; Georges, G.; Shi, J.; Deane, C. M. “SAbDab: The Structural Antibody Database”, Nucleic Acids Res 2014, 42 (D1). https://doi.org/10.1093/nar/gkt1043.
- Kiyoshi, M.; Caaveiro, J. M. M.; Miura, E.; Nagatoishi, S.; Nakakido, M.; Soga, S.; Shirai, H.; Kawabata, S.; Tsumoto, K. “Affinity Improvement of a Therapeutic Antibody by Structure-Based Computational Design: Generation of Electrostatic Interactions in the Transition State Stabilizes the Antibody-Antigen Complex”, PLoS One 2014, 9 (1). https://doi.org/10.1371/journal.pone.0087099.


