Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Cambridge, UK – 14 July 2021 – Cresset, innovative provider of outstanding software for molecule discovery and design, announces the release of Flare V5, its comprehensive molecule design platform.
This release of Flare incorporates the advanced ligand-based methods validated through years of use in Forge™ with its established structure-based science. The integration enables chemists to discover novel molecules more efficiently and effectively. Significant enhancements have also been made to increase the speed and automation of Free Energy Perturbation (FEP) calculations, enabling single compounds to be predicted in as little as 30 minutes. New analysis methods have been added to help understand the behavior of water within the target active site.
“Flare has evolved from a structure-based design tool” says Dr Giovanna Tedesco, Head of Products. “In this exciting development, ligand and structure-based methods work in full synergy, allowing medicinal and computational chemists to access Cresset intuitive science in a single platform to advance their drug discovery projects.”
“Flare V5 also features new and enhanced science, and improvements to the GUI to increase usability. We now offer a comprehensive solution to streamline molecule design and the prioritization of new molecules for synthesis.”
Flare is available for evaluation upon request.
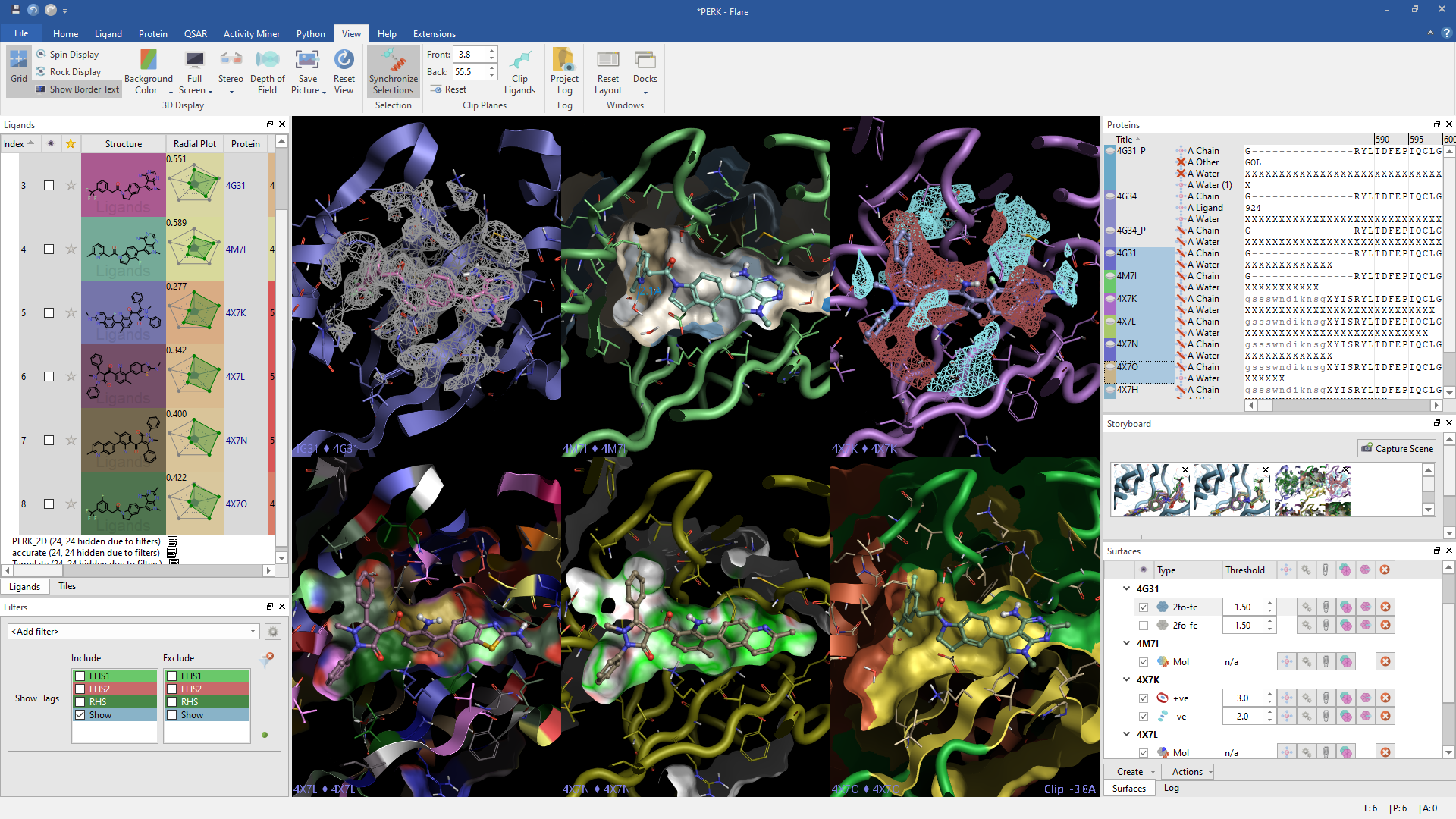
Flare enables research chemists to discover novel small molecules more efficiently and effectively in a single platform