Introduction
Fragment-based drug discovery (FBDD) is well-established as an effective method for advancing innovative drug design and has been adopted as a valuable technique for developing novel, diverse hits. FBDD has contributed to the development of several approved and clinical-phase drugs, including Vemurafenib, Venetoclax, Pexidartnib and Onalespib.
A challenge of fragment-based drug discovery is being able to morph experimentally-identified (by isothermal titration calorimetry (ITC), NMR, SPR, MS and X-ray crystallography) small, weak affinity binding fragments into suitable ligands either through merging, linking, or growing. This is where medicinal chemistry experience and computational expertise play an essential role, as well as having access to specialist software solutions. Cresset Discovery’s expert modelers apply their deep computational knowledge and experience to explore and optimize bioisosteric replacements to prioritize starting lead compounds using SparkTM 1 one of the best independently validated software tools for linking, growing and merging fragments2.
Identifying the fragment binding site in the protein target
When fragments have been identified through NMR and X-Ray methods, the protein target binding pockets are known. However, if fragments are identified through SPR and ITC, the fragment binding site may need to be identified. Cresset Discovery’s consultants apply pocket detection for identification and characterization of potential druggable binding sites3.
An example of designing PNMT inhibitors using fragment-based drug discovery
Phenylethanolamine N-methyltransferase (PNMT) participates in the synthesis of adrenaline (epinephrine) in the central nervous system (CNS) by catalyzing the methylation of noradrenaline. Therapeutics targeting PNMT are desired for treatment of many pathologies including blood pressure control, Parkinson’s disease, Alzheimer’s disease, and pituitary hormone secretion abnormalities.
It is a challenging drug target since the solved crystal structure with substrates/inhibitors shows a closed active site (PDB:1HNN, 3HCD) (Figure 1A) and no apo-crystal structures are available. Additionally, the enzyme cannot be crystallized in the absence of s-adenosyl-l-homocysteine (AdoHcy) which is present in the cofactor binding site in the crystal structure. X-ray structures of PNMT solved with fragments (PDB codes: 3KPJ, 3KPU, 3KPV, 3KPW, 3KPY, 3KQM, 3KQS, 3KQT, 3KQV, 3KQW, 3KQO, 3KQP, 3KQQ, 3KQY, 3KR0, 3KR1 and 3KR2) (example shown in Figure 2A) identified4 from the rapid crystal structure soaking method served as the basis for illustrating a fragment-based drug design approach as would typically be performed by Cresset Discovery’s dedicated team of CADD scientists.
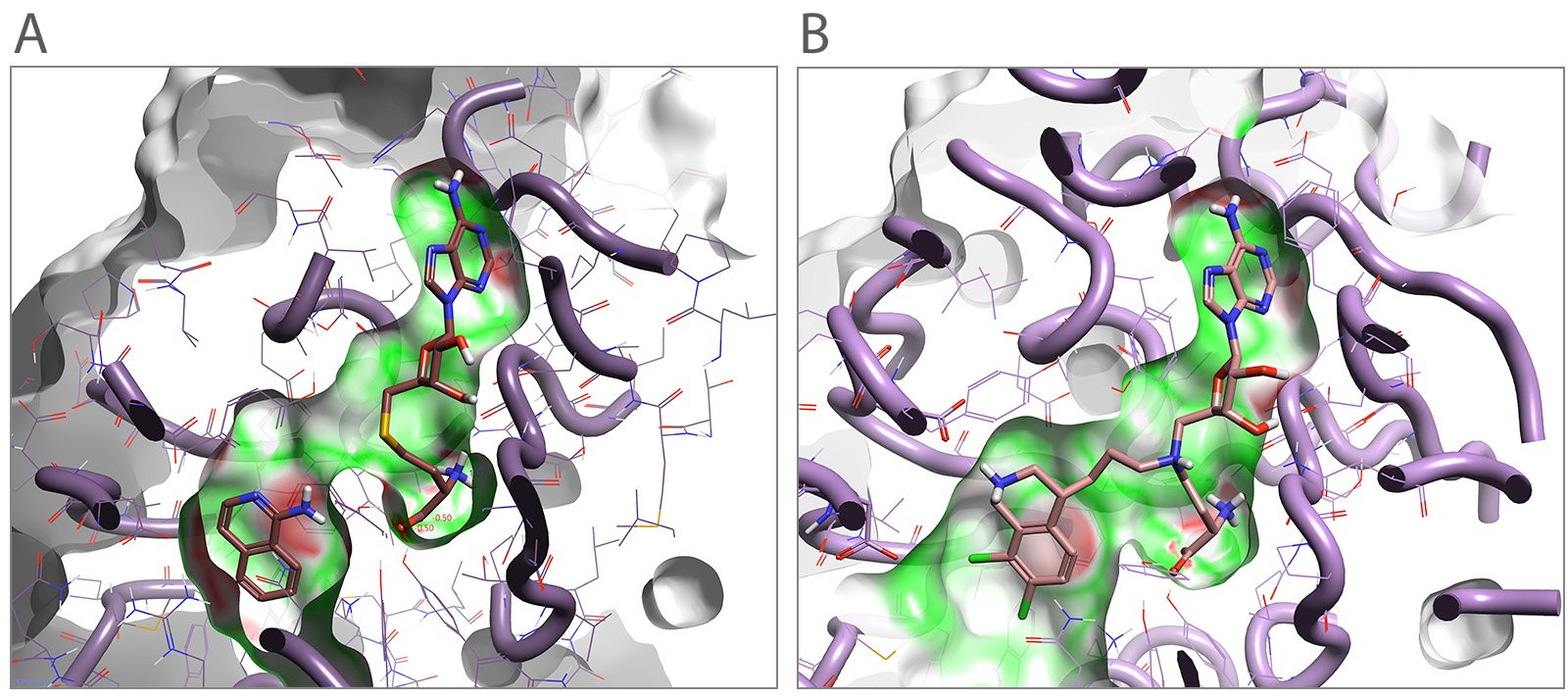
Figure 1. (A) Human PNMT crystal structure is shown with a fragment crystallized together with s-adenosyl-l-homocysteine (AdoHcy). (B) Illustration of how fragments can be linked in this binding pocket. The protein binding pocket is colored illustrating Electrostatic Complementarity™ to the protein (green illustrating good complementarity and red poor complementarity)
In the literature study, a fragment-based screening library was used to identify and crystalize fragments. Fragments in PNMT were crystallized with s-adenosyl-l-homocysteine (AdoHcy, Figure 1A) which binds to the cofactor site and is required for crystallization. These small library crystallized fragments displace a phosphate found in the noradrenaline binding pocket.
Cresset Discovery scientists applied de novo design expertise, in conjunction with analysis using bioisosteric replacement1 and modeling3, to grow these small crystal structure fragments, evaluate their Electrostatic Complementarity to the protein, and analyze protein-ligands interactions within the protein binding site. Additionally, fragments could be linked with designed s-adenosyl-l-homocysteine linkers as a starting point for ligand design to fill the complete ligand-cofactor binding pocket.
![]()
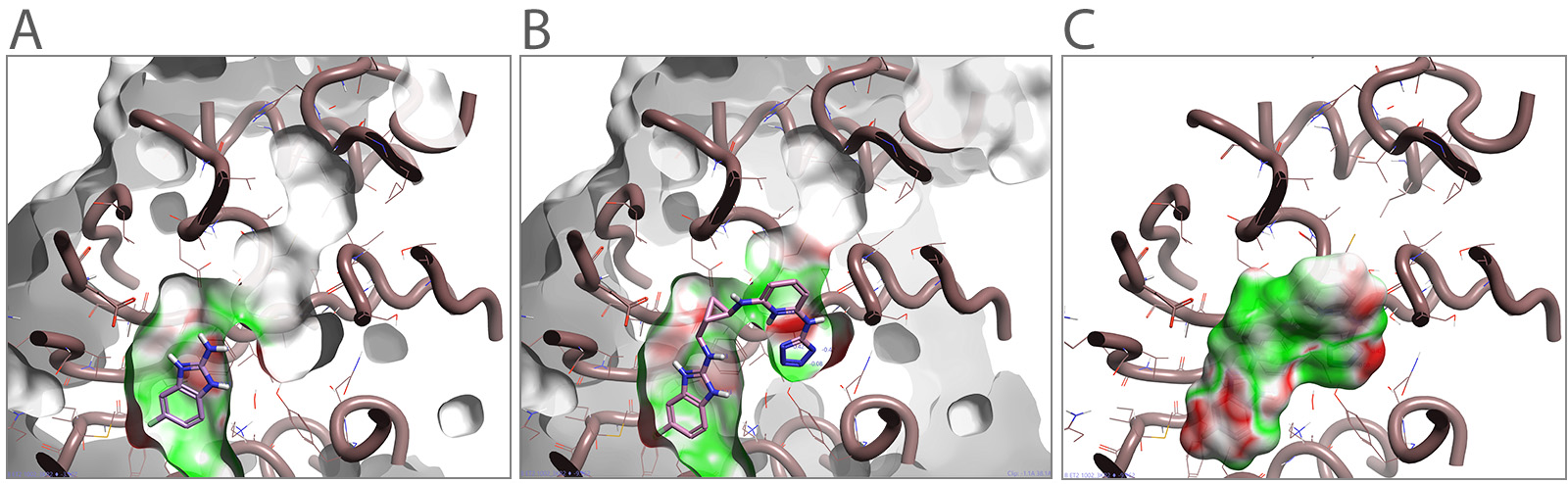
Figure 2. (A) An example of PNMT with a crystallized fragment. (B) PNMT fragment grown in the binding pocket using s-adenosyl-l-homocysteine as a growing guide (C) Designed ligand colored according to the Electrostatic Complementarity to the protein structure.
Beginning with a starter ligand, and modeling against one or more reference molecules (AdoHcy in this case), Cresset Discovery scientists mapped a different region of the same pocket within the protein. The starter and reference molecules are both pre-aligned with respect to each other in 3D. Ligand joining experiments could also be performed using the crystalized fragment and AdoHcy as two starter molecules which can then be linked with different suggested linkages. This generates valuable starting leads to accelerate generating IP with novel FBDD ligands.
In addition to ligand growing and joining, Cresset Discovery can provide specialist solutions to facilitate ligand macrocyclization, water replacement or even generate fragment databases.
Engage Cresset Discovery to advance your FBDD project
As your premier computational chemistry CRO, we have the required skillset and resources to support your fragment-based discovery project by growing and linking fragments, facilitating de novo ligand design and increasing the diversity of your ligand modifications. To discover how we can work alongside your team, request a free confidential discussion.
References
- Spark™, Cresset®, Litlington, Cambridgeshire, UK; https://www.cresset-group.com/spark/; Cheeseright T., Mackey M., Rose S., Vinter, A.; Molecular Field Extrema as Descriptors of Biological Activity: Definition and Validation. J. Chem. Inf. Model. 2006, 46 (2), 665-676
- Matthew P. Baumgartner and David A. Evans, Side chain virtual screening of matched molecular pairs: a PDB-wide and ChEMBL-wide analysis, Journal of Computer-Aided Molecular Design 2020, 34, 953–963.
https://doi.org/10.1007/s10822-020-00313-1
- Flare™, Cresset®, Litlington, Cambridgeshire, UK; https://www.cresset-group.com/flare/; Cheeseright T., Mackey M., Rose S., Vinter, A.; Molecular Field Extrema as Descriptors of Biological Activity: Definition and Validation J. Chem. Inf. Model. 2006, 46 (2), 665-676; Bauer M. R., Mackey M. D.
- Nyssa Drinkwater, Hoan Vu, Kimberly M. Lovell, Kevin R. Criscione, Brett M. Collins, Thomas E. Prisinzano, Sally-Ann Poulsen, Michael J. Mcleish, Gary L. Grunewald and Jennifer L. Martin, Biochem. J. 2010, 431, 51-61. https://doi.org/10.1042/BJ20100651

