Antibodies are fantastically versatile molecular recognition engines, capable of creating artificial enzymes in response to recognized interactions. Similarly, enzymes depend on molecular recognition for catalysis. Cresset Discovery Services has a depth of experience in modeling molecular recognition scenarios, stretching back to the early nineties.
Nature’s molecular recognition engines
In 1992 I was about to start a PhD on ‘catalytic antibodies’, investigating the amazingly versatile ability of the immune system to prepare artificial enzymes. It all sounded fascinating, and still does. Alas, rather than building antibodies, the path of my PhD altered, resulting in me developing my organo-phosphorus chemistry skills and producing ‘transition-state inhibitors’ of beta lactamase enzymes instead.
However, I retained a keen interest in molecular recognition and a familiarity with antibodies as a protein class and this has proved useful for our work at Cresset Discovery Services today. Indeed, it wasn’t until much, much, later that immunoglobulins would reveal their impact as important future medicines and of course this was way before the multi-billion-dollar blockbuster TNFa targeting biologic Adalibumab (Humira) (Figure 1, right).
Immunoglobulins are biologically evolved to recognize an enormously variable patch of molecular surface using a single protein architecture; an arrangement of two polypeptide chains (heavy and light) each bearing three hyper variable loops.
Figure 1 illustrates the diversity of recognition possible in antibodies, as modeled in Flare™, ranging from small molecules (sulfathiazole) to big molecules (Fullerene C60)1 and peptides (TNFa)2.
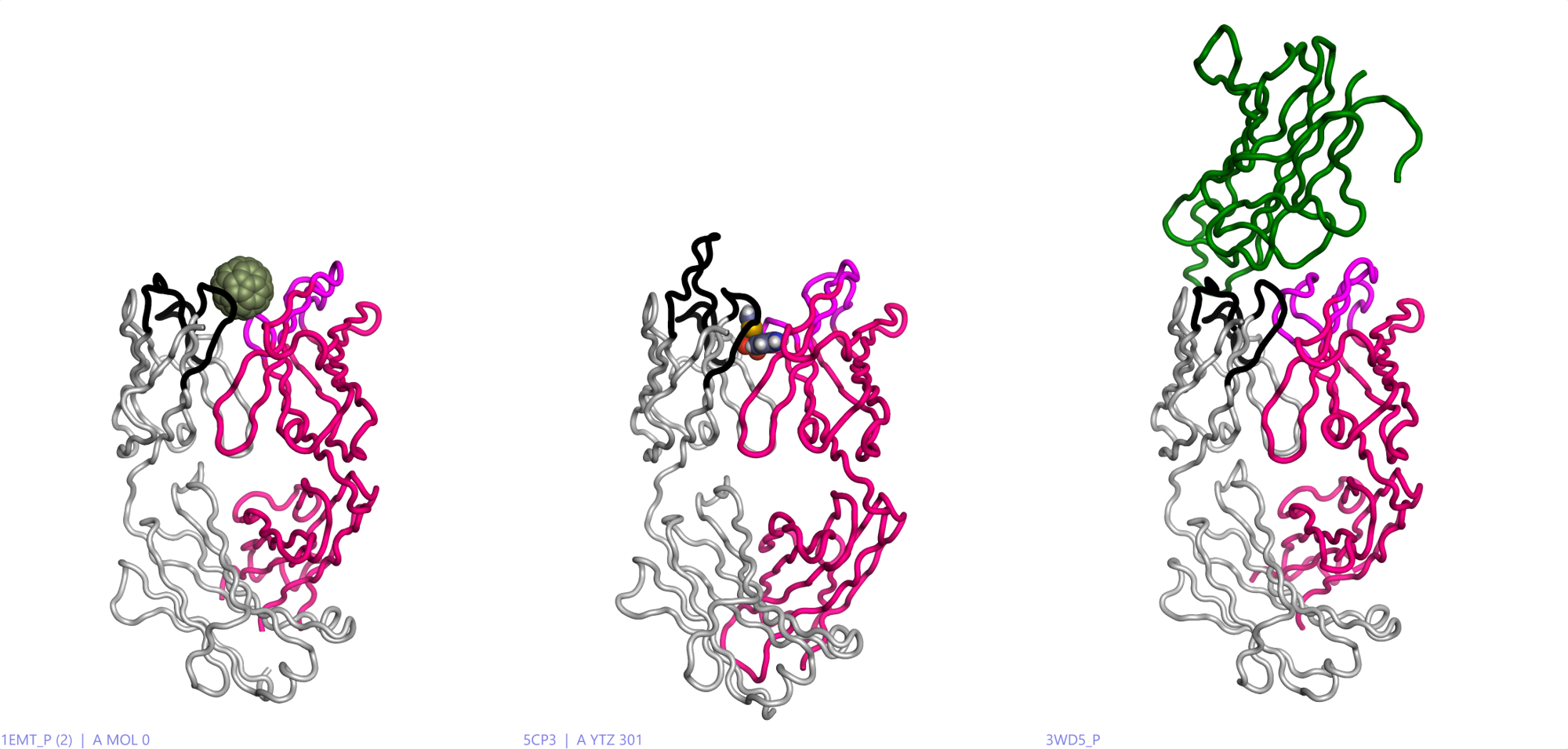
Figure 1: Left: an example of a fullerene recognizing mouse antibody. Middle:another mouse antibody recognizing a sulphonamide drug, Sulfathiazole. Right: the human engineered antibody Adalibumab (green) with TNFa. Heavy chain (crimson) variable loops (magenta), Light chain (grey) variable loops (black). All modeled in Flare using PDB codes: 5CP3, 1EMT and 3WD5.
Similarly, enzymes also use molecular recognition to great advantage via a diverse array of protein architectures. In contrast to antibodies, they specifically recognize molecular surfaces that represent a transition state of a chemical transformation. Lowering the energy of the transition state through binding allows catalysis to happen in enzymes.Figure 1: Left: an example of a fullerene recognizing mouse antibody. Middle:another mouse antibody recognizing a sulphonamide drug, Sulfathiazole. Right: the human engineered antibody Adalibumab (green) with TNFa. Heavy chain (crimson) variable loops (magenta), Light chain (grey) variable loops (black). All modeled in Flare using PDB codes: 5CP3, 1EMT and 3WD5.
The main premise of catalytic antibodies is to elicit their production in response to a transition state that can be mimicked by a ligand structure, which in theory then reproduces enzymic behaviour in the resultant protein. This pursuit simultaneously probes our fundamental knowledge of molecular recognition and of enzyme catalysis.
A catalytic antibody that cleaves cocaine
In 2006 workers at Scripps published work on a catalytic antibody3 that cleaves cocaine at the benzoyl ester. The antibody was elicited using an aryl phosphonate ester – a mimic of the carboxyl ester hydrolysis transition state.
The insights gained through the X-ray structures that were solved for this system were remarkable. Multiple components of the reaction coordinate are shown to exploit common recognition patterns, whilst the different geometries are accommodated by both residue and backbone movements in a dynamic process (Arg and Tyr movements in Figure 2).
This is an excellent model system. It shows how some interactions are very favourable e.g., the cation-pi interaction of the tropane is very solid across the structures, whilst others e.g., the ester, are variable. It also demonstrates that proteins, and in particular enzymes, ‘breathe’ – they are not statues (Figure 2).
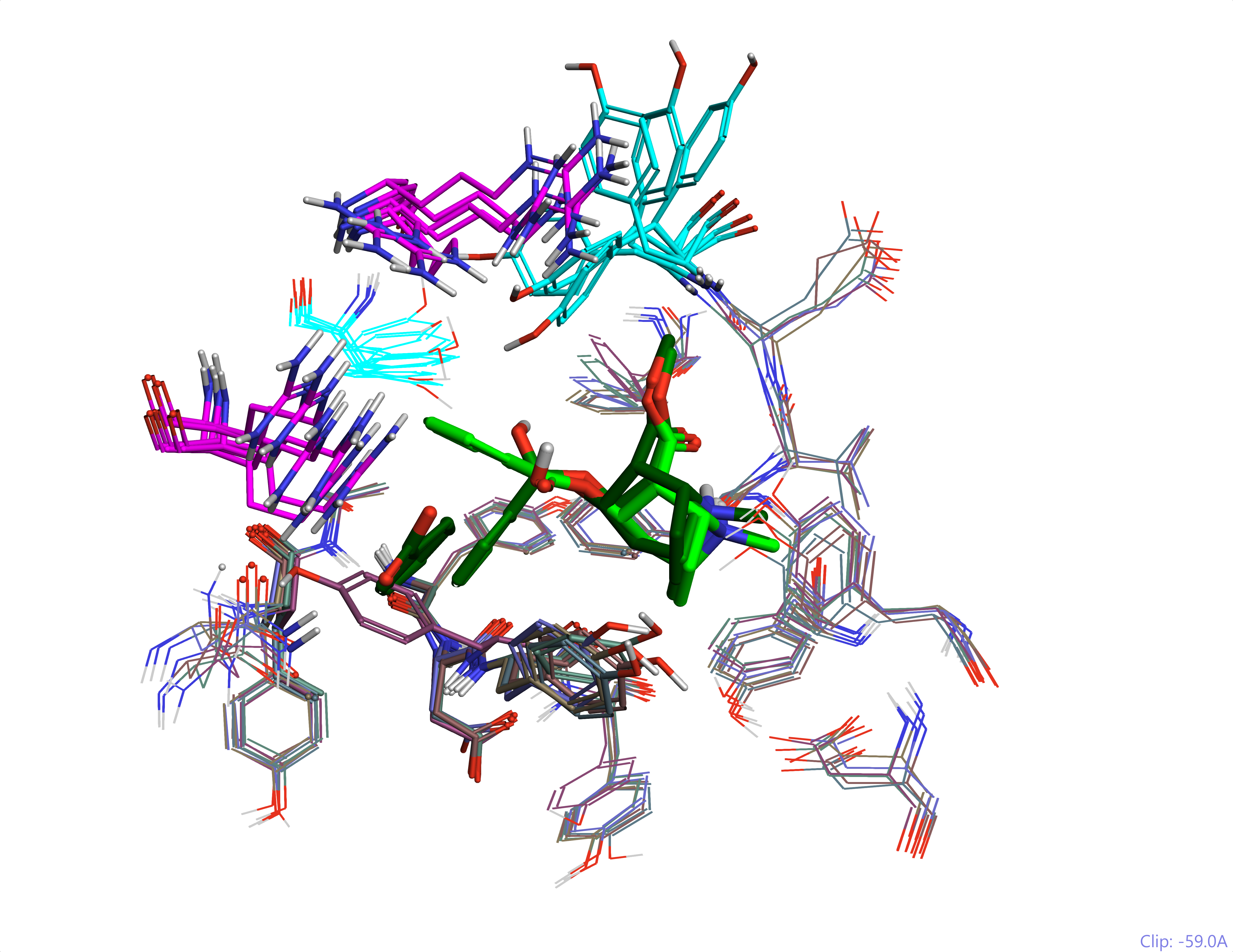
Figure 2: The Scripps catalytic antibody in the different protein conformations that recognize the substrate and the transition state for cocaine ester hydrolysis. This shows highly mobile Arg (magenta) and Tyr (cyan) residues and the ligand (cocaine hydrolysis reaction coordinate – light to dark green). All modeled in Flare using PDB codes: 2AJU, 2AJV, 2AJZ, 2AJX, 2AJY and 2AJS.
All molecular recognition is not equal
One very interesting facet of molecular recognition demonstrated by antibody antigen interactions is that these interactions are not all equal. In fact, there are lesser and greater interactions – the latter of which can predominate in driving antibody production. Preferred interactions (or ‘hot spots’) can be visualised using various techniques (e.g., as demonstrated using the previous cocaine system modeling in Flare shown in Figure 3).
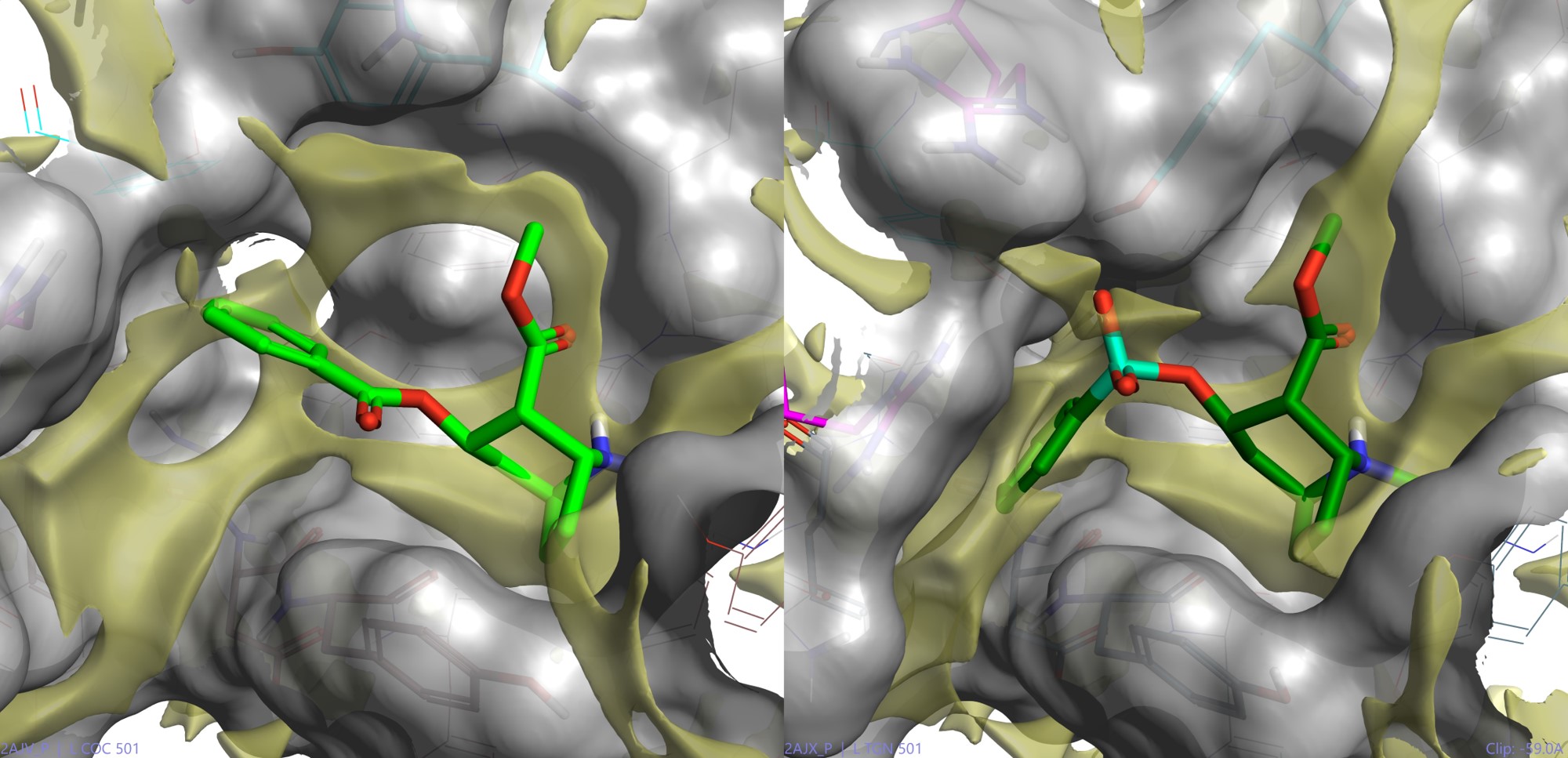
Figure 3: The Scripps catalytic antibody conformer ligand interaction surfaces. All modeled in Flare using PDB codes: 2AJV and 2AJX. This shows highly mobile surface changes required to squash the substrate into the transition state geometry (this time with the phosphono mimetic) and the hydrophobic hot spot surface (yellow) calculated by Flare.
Antibodies are produced via natural selection processes as a cellular response to a presented antigen. Some parts of the antigen are better than others at eliciting antibody responses, since they may involve preferred interactions, and so the resultant antibody may not bind in a therapeutically useful way.
Modeling and design of these interactions (i.e., via antigen engineering) becomes a useful task, since leaving it to nature can divert us from our intended goals.
Cresset Discovery Services work on biologics
While we are actively working and delivering on projects involving biologics, client confidentiality means that it remains a challenge to describe details of work done for commercial clients. We can, however, speak more generally and we have recently delivered on projects to:
- Predict / design mutants that prevent binding
- Model strategic glycan positioning at unhelpful yet potently antigenic surfaces
- Model / characterize the observed binding order of an antigen – receptor series.
Read more about Cresset’s capabilities in biologics: Modeling ‘big’: Applying the XED force field to biologics.
Biologics remain a very important class of protein targets for disease therapeutics which we intend to continue to support through our innovative modeling services.
Free confidential discussion
Find out how we can accelerate your project by requesting a free confidential discussion.
References
- X-ray crystal structure of an anti-Buckminsterfullerene antibody Fab fragment: Biomolecular recognition of C60, Braden*, B. C., Goldbaum F. A., Chen B., Kirschner A. N., Wilson S. R. and Erlanger B. F. PNAS 97, no. 22, 12193–12197, 2000.
- Comparison of the Inhibition Mechanisms of Adalimumab and Infliximab in Treating Tumor Necrosis Factor a-Associated Diseases from a Molecular View, Shi Hu, Shuaiyi Liang, Huaizu Guo, Dapeng Zhang, Hui Li, Xiaoze Wang, Weili Yang, Weizhu Qian, Sheng Hou, Hao Wang§, Yajun Guo and Zhiyong Lou, Journal of Biological Chemistry, 288, 38, pp. 27059 –27067, 2013
- Complete Reaction Cycle of a Cocaine Catalytic Antibody at Atomic Resolution, Zhu X., Dickerson T. J., Rogers C. J., Kaufmann G. F., Mee J. M., McKenzie K. M., Janda K. D. and Wilson I. A. Structure 14, 205–216, 2006.


