Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Docking is a method that utilizes protein structures to generate ligand binding poses and scores the interactions that are made. This method is used extensively in drug discovery to screen molecule designs and help prioritize them. Docking software typically treats the protein as a rigid body, whilst the ligand is conformationally flexible. Occasionally this is problematic because the protein structure is not in the biologically relevant conformation for a given ligand, which leads to the docking results not being relevant. In this case study, we will firstly validate the approach of using molecular dynamics (MD) to produce an ensemble of protein conformations to be used with ensemble docking in Flare.
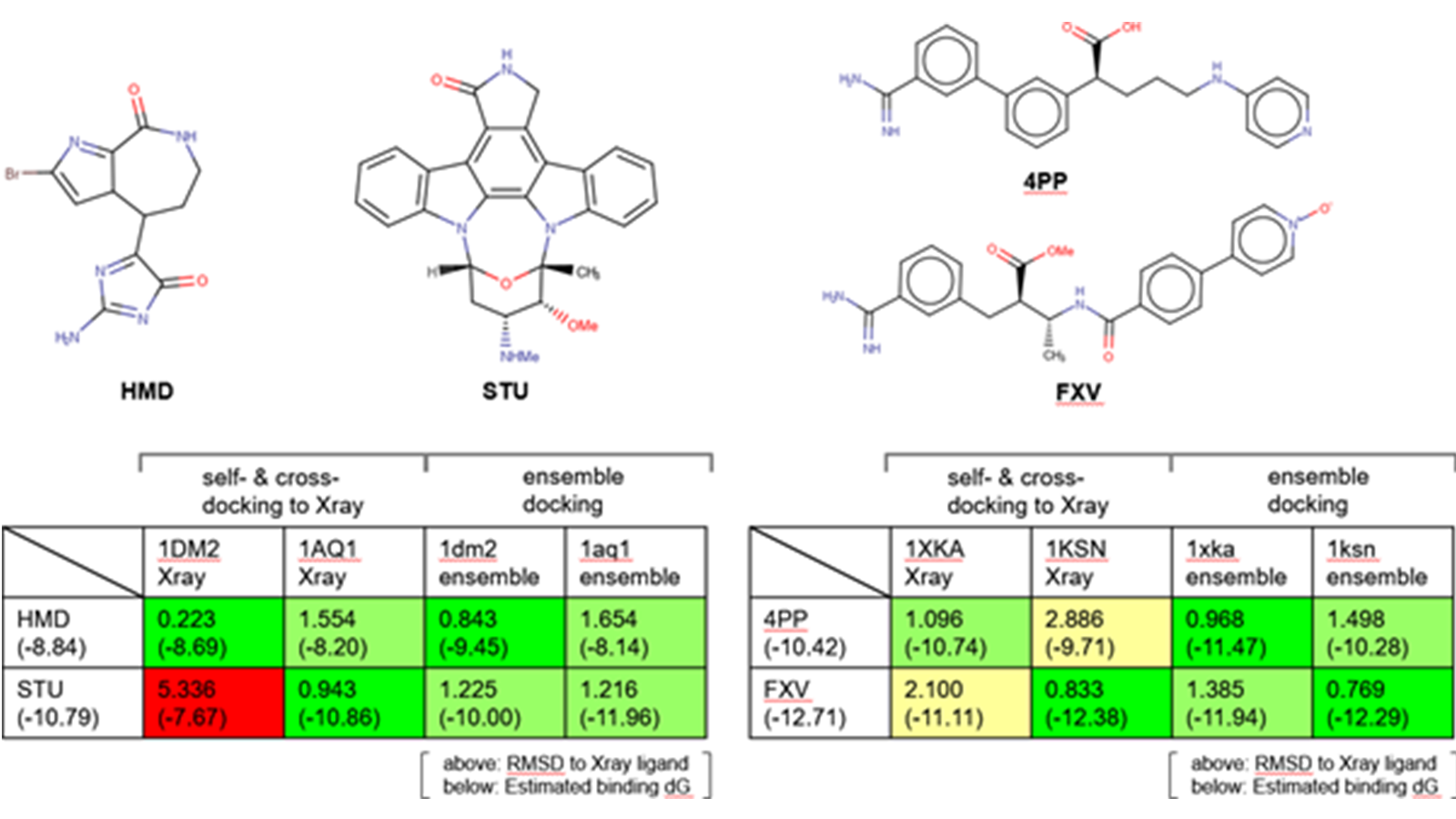
Verification of MD/ensemble docking. Left: self-docking and cross-docking results for CDK2 ligands. Right: self-docking and cross-docking results for Factor Xa ligands.