Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
To accompany the release of Spark™ V10.6, the Spark fragment and reagent databases have been updated and are now available for download. Derived by fragmenting compounds and reagents from commercial sources and the literature, these database are a great source of novel ideas for your drug discovery projects, ensuring at the same time that the results found by Spark are always associated to real, synthetically accessible compounds.
The Spark ‘Commercial’ databases in this release are derived from the eMolecules Screening Compounds. With more than 6 million fragments to search overall, they provide an excellent source of chemical diversity for your experiments.
The Spark ‘ChEMBL’ databases have also been updated. Based on release 26 of ChEMBL, they provide more than 1.5 million additional fragments to search, derived from chemical literature compounds.
Compounds in both original source collections are filtered to remove molecules containing potentially toxic or reactive groups before the creation of the databases. Each compound is then fragmented independently, breaking the bonds which connect to heteroatoms, carbonyls, thiocarbonyls and bonds to rings. Specific functional groups such as carboxylic acids, nitro groups and rings are not fragmented. The frequency with which a given fragment occurs is captured together with the number of bonds that were broken to disconnect the fragment from the parent molecule.
All resultant fragments are subject to molecular weight, number of H-bond acceptor/donor and rotatable bond limits. They are then sorted by frequency and labelled as shown in Table 1.
Table 1: Fragment databases sorted by frequency.
| Spark category | Database | Total number of fragments (to nearest 1,000) | Frequency |
| Commercial | Very Common | 68,000 | Fragments which appear in more than 725 molecules |
| Common | 68,000 | Fragments which appear in 215-724 molecules | |
| Less Common | 212,000 | Fragments which appear in 65-214 molecules | |
| Rare | 280,000 | Fragments which appear in 25-64 molecules | |
| Very Rare | 527,000 | Fragments which appear in 9-24 molecules | |
| Extremely Rare | 534,000 | Fragments which appear in 5-8 molecules | |
| Ultra Rare | 770,000 | Fragments which appear in 3-4 molecules | |
| Doubleton* | 1,053,000 | Fragments which appear in 2 molecules | |
| Singleton* | 2,526,000 | Fragments which appear in a single molecule | |
| ChEMBL | Common | 232,000 | Fragments which appear in more than 12 molecules |
| Rare | 232,000 | Fragments which appear in 4-12 molecules | |
| Very Rare | 382,000 | Fragments which appear in 2-3 molecules | |
| Extremely Rare* | 641,000 | Fragments which appear in a single molecule |
*Contact us for further details.
Typically we would recommend to install only the databases including fragments which appear at least 3-4 times in the original collections. The databases containing fragments seen with lower frequency (Singleton, Doubleton and ChEMBL Extremely Rare) are very large, and may contain fragments derived from unrealistic/wrong structures in the original collections. If you do wish to use these databases then please contact Cresset Support for download instructions.
The number of fragments in each database per connection point count (excluding the databases containing only singletons and doubletons) is shown in Figure 1.
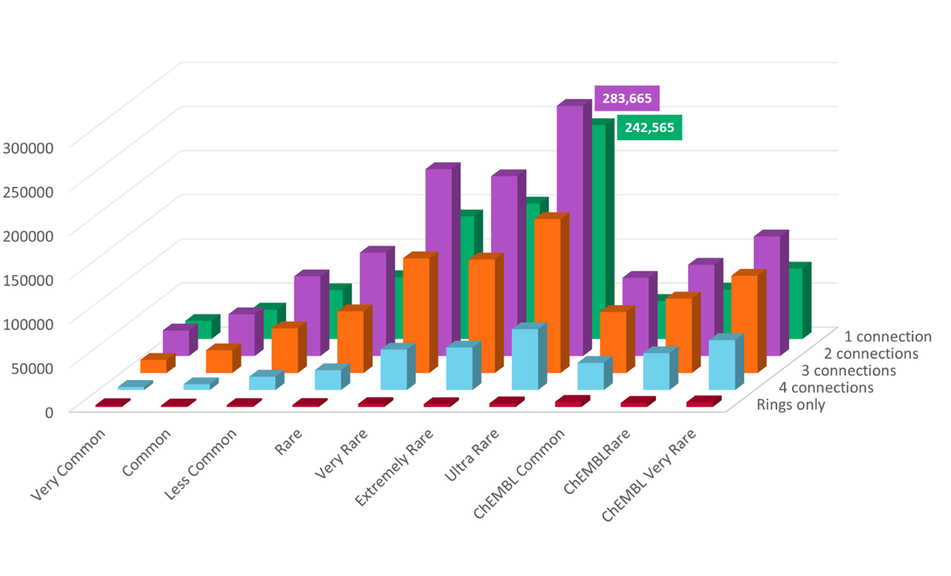
Figure 1: Count of fragments in Spark ‘Commercial’ and ‘ChEMBL’ databases split by the number of connection points of each fragment.
The most common fragments in the ChEMBL and Commercial databases have a significant overlap (Table 2). However, comparing the rarer fragments from each database shows significantly less overlap, highlighting the different areas of chemical space each database occupies.
Table 2: Overlap of the most common fragments in the ChEMBL and Commercial databases.
| % overlap | Very Common | Common | Less Common | Rare | Very Rare | Extremely Rare | Ultra Rare | Doubleton* | Singleton* | Unique |
| ChEMBL common | 17% | 14% | 13% | 8% | 8% | 4% | 4% | 3% | 5% | 24% |
| ChEMBL rare | 2% | 6% | 9% | 9% | 10% | 6% | 5% | 4% | 6% | 43% |
| ChEMBL very rare | 1% | 2% | 4% | 5% | 7% | 5% | 5% | 5% | 7% | 58% |
| ChEMBL extremely rare* | 0% | 1% | 2% | 3% | 5% | 4% | 4% | 4% | 9% | 68% |
*Contact us for further details.
With more than 6.9 million unique fragments to search, the Spark fragment databases provide an extremely large source of novel bioisosteres for Spark projects, which can be further complemented by generating fragments from your corporate collection with the Spark Database Generator, a dedicated and user-friendly interface to custom database creation within Spark.
Monthly updates of the Spark reagent databases, derived from the eMolecules building blocks using an enhanced set of rules for chemical transformation, are included in the Spark V10.6 release. The November update includes over 314,000 reagents with up-to-date availability information, to make it easy for you to order the reagents you require to synthesize your favorite Spark results.
| Total | 1-50 | 51-100 | 101-150 | 151-200 | 201-250 | |
| eMolecules_acidCO | 23,983 | 3 | 401 | 6,732 | 13,361 | 3,486 |
| eMolecules_acid | 41,545 | 43 | 2,811 | 15,618 | 17,939 | 5,134 |
| eMolecules_alcohol | 18,032 | 11 | 1,435 | 7,521 | 7,193 | 1,872 |
| eMolecules_alcoholO | 19,634 | 3 | 468 | 6,773 | 9,666 | 2,724 |
| eMolecules_aliphatic_halide | 8,808 | 13 | 924 | 3,651 | 3,421 | 799 |
| eMolecules_alkyne | 2,851 | 27 | 505 | 1,420 | 781 | 118 |
| eMolecules_aromatic_alcoholO | 8,625 | 0 | 44 | 1,927 | 5,023 | 1,631 |
| eMolecules_aromatic_aminesN | 18,557 | 0 | 111 | 4,207 | 10,567 | 3,672 |
| eMolecules_aromatic_halide | 40,110 | 8 | 451 | 13,592 | 22,762 | 3,297 |
| eMolecules_boronic | 4,496 | 0 | 128 | 1,894 | 2,093 | 381 |
| eMolecules_cyano | 15,118 | 20 | 1,086 | 5,662 | 6,283 | 2,067 |
| eMolecules_isocyanateCO | 555 | 0 | 20 | 170 | 287 | 78 |
| eMolecules_olefin | 3,273 | 16 | 524 | 1,419 | 1,089 | 225 |
| eMolecules_primary_aliphatic_amine | 19,016 | 6 | 1,366 | 8,495 | 7,763 | 1,386 |
| eMolecules_primary_aliphatic_amineN | 11,571 | 0 | 398 | 5,234 | 5,101 | 838 |
| eMolecules_primary_aliphatic_halide | 6,875 | 12 | 627 | 2,886 | 2,705 | 645 |
| eMolecules_primary_aromatic_amines | 23,350 | 0 | 325 | 6,581 | 12,171 | 4,273 |
| eMolecules_reductive_amination | 22,127 | 3 | 818 | 6,551 | 10,683 | 4,072 |
| eMolecules_secondary_aliphatic_amineN | 15,061 | 1 | 277 | 4,270 | 8,413 | 2,100 |
| eMolecules_sulfonicacid | 5,066 | 31 | 602 | 2,265 | 1,761 | 407 |
| eMolecules_sulfonicacidSO2 | 3,075 | 0 | 13 | 302 | 1,584 | 1,176 |
| eMolecules_thiol | 721 | 7 | 206 | 330 | 164 | 14 |
| eMolecules_thiolS | 1,986 | 1 | 38 | 537 | 1,078 | 332 |
In the Spark results table, the eMolecules IDs for your favorite reagents can be easily exported from Spark and used to purchase the compounds from the eMolecules building blocks database, as shown in the web clip How to use the eMolecules reagents databases in Spark and access ordering information for the result.
Spark V10.6 also includes the new ‘COD’ database (Figure 2). This contains more than 440K fragments in their crystallographic conformation, derived from the Crystallography Open Database and available for download to all Spark customers.
Figure 2: The new ‘COD’ database is available to all Spark customers and includes more than 440K fragments in their crystallographic conformation.
If you have access to large collections of proprietary chemistry or specialized reagents, or if you want to only consider fragments from reagents you have in stock, you can add value to your Spark experiments by creating your own custom databases.
These can be easily prepared using the Database Generator (Figure 3), a dedicated and user-friendly interface to custom database creation within Spark, or using the equivalent functionality from the command line.
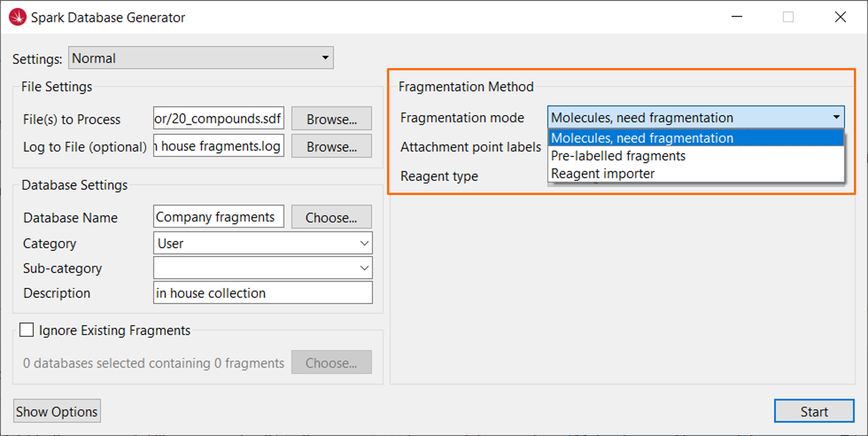
Figure 3: Use the Spark Database Generator to create your own fragments and reagent databases.
This new release of the fragment and reagent databases, combined with custom databases from corporate collections generated with the Spark Database Generator, will provide an outstanding range of bioisosteres for your project.
Please contact us to update to the latest databases, to learn how to make the best use of the Spark Database Generator, or to find out how Spark can impact your project.