Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Unlike existing methods, PickR™ V1.0 uses the 3D electrostatic and shape properties of molecules to cluster and select R-groups for inclusion in hit-finding and hit-to-lead libraries. This command line application is easy to use, interfaces to most common queuing systems like Grid Engine, PBS or LSF, and generates logical associations between molecules that fits with the expectations of both medicinal and computational chemists. 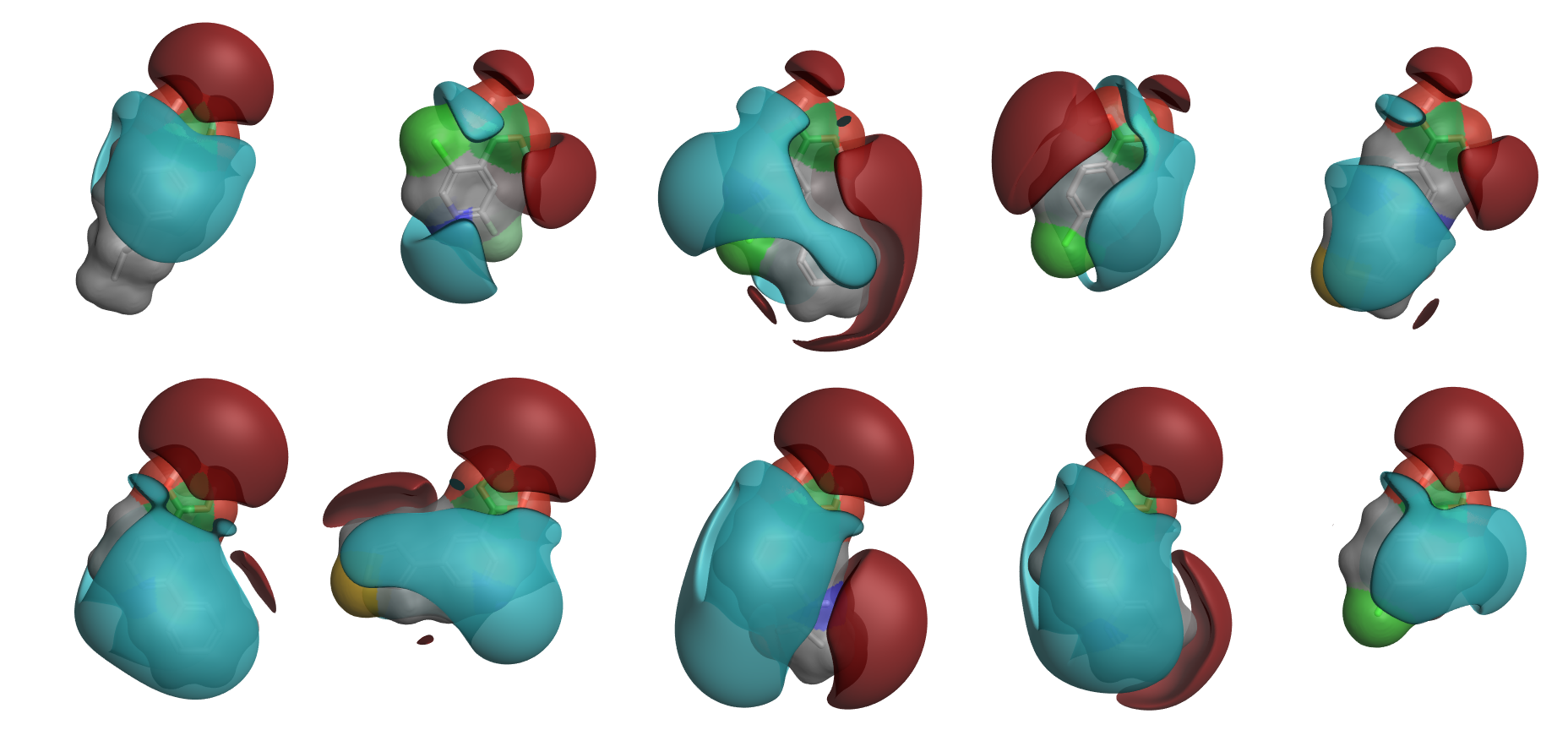
Figure 1: Medoids of the largest 10 clusters from running PickR on 5,500 boronic acids. Positive interaction potential: Red; Negative potential: Blue.
The PickR algorithm utilizes the concept that most libraries are constructed using a combinatorial paradigm, such that the selection of the final molecules to be included in the library can be simplified to the selection of a suitable range of building blocks, or R-groups. To assess the diversity of these R-groups, we align all reagents on a common bond, usually the bond formed in the combinatorial reaction, and compute the electrostatic and shape similarity of every pair of conformations. As the alignment along a bond involves a rotational degree of freedom, we sample multiple mutual arrangements of each reagent pair to make sure that the best steric and electrostatic overlap is attained.
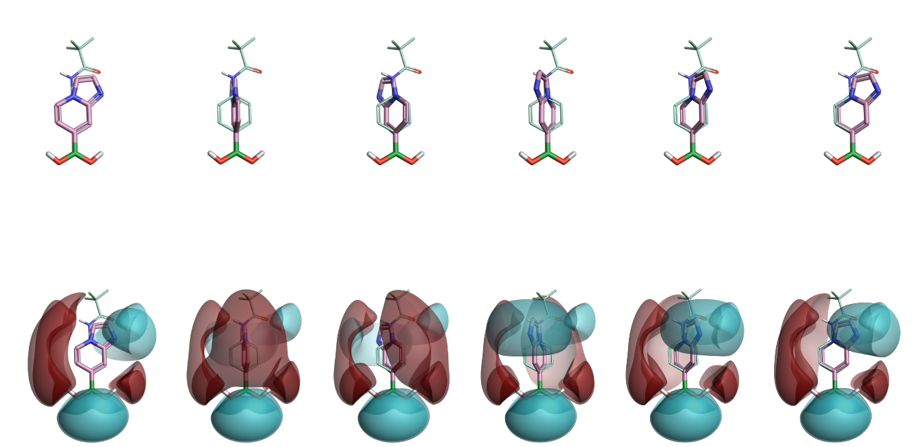
Figure 2. Alignment and rotation along the boron-carbon bond and the generated electrostatic map for two boronic acids. Repetition across conformations leads to a single similarity value for the pair.
Using PickR results in different similarities for reagents over the traditional 2D fingerprint methodologies. The electrostatic and shape generates both expected and unexpected relationships that are poorly described using 2D measures. The difference in 3D is particularly acute when studying small highly functionalized rings which form many of the common reagents in modern library design. However, 2D similarity is less sensitive to regiochemistry than 3D and hence we recommend using a combination is most practical experiments.
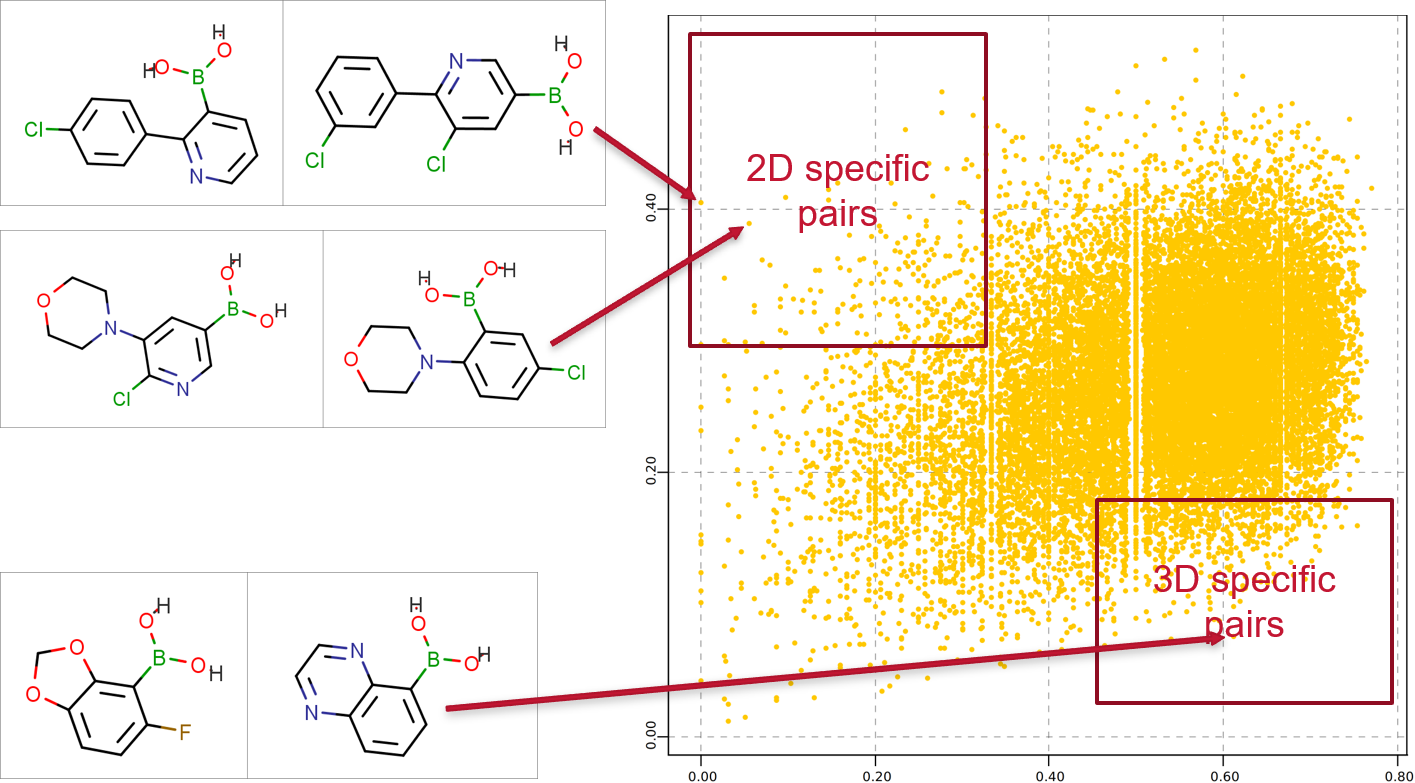
Figure 3: A comparison of 2D and 3D distance scores obtained for pairs of molecules in a random selection of 200 commercially available boronic acids using PickR and RDKit (MACCS) fingerprints.
Using a 3D electrostatic and shape similarity measure across molecule conformations is computationally more demanding than the using 2D fingerprints. PickR is designed to move the computational process to a Linux cluster with minimal effort. This is achieved through support for most popular queuing engines (SGE, PBS, :LSF) and automated job submission. A simple command line switch is used to specify the cluster submission enabling processing of many thousands of reagents in a reasonable time.
The utility of PickR is visible from the results of a clustering experiment on a large collection of commercially available boronic acids. The reagents were download from eMolecules’ database using atom count and rotatable bond limits (#Atoms<18, rotatable bond=3 atoms). PickR was applied using the default options as follows
pickr -s '[B:1][#6:2]’ \ #bond to break specified as SMARTS -Q sge \ #use SGE queuing -j 200 \ #use a max 200 jobs -v \ #verbose output boronics.smi #smiles file (sdf also)
The calculation took a few hours on 150 cpu cores and generated 550 clusters (the default).
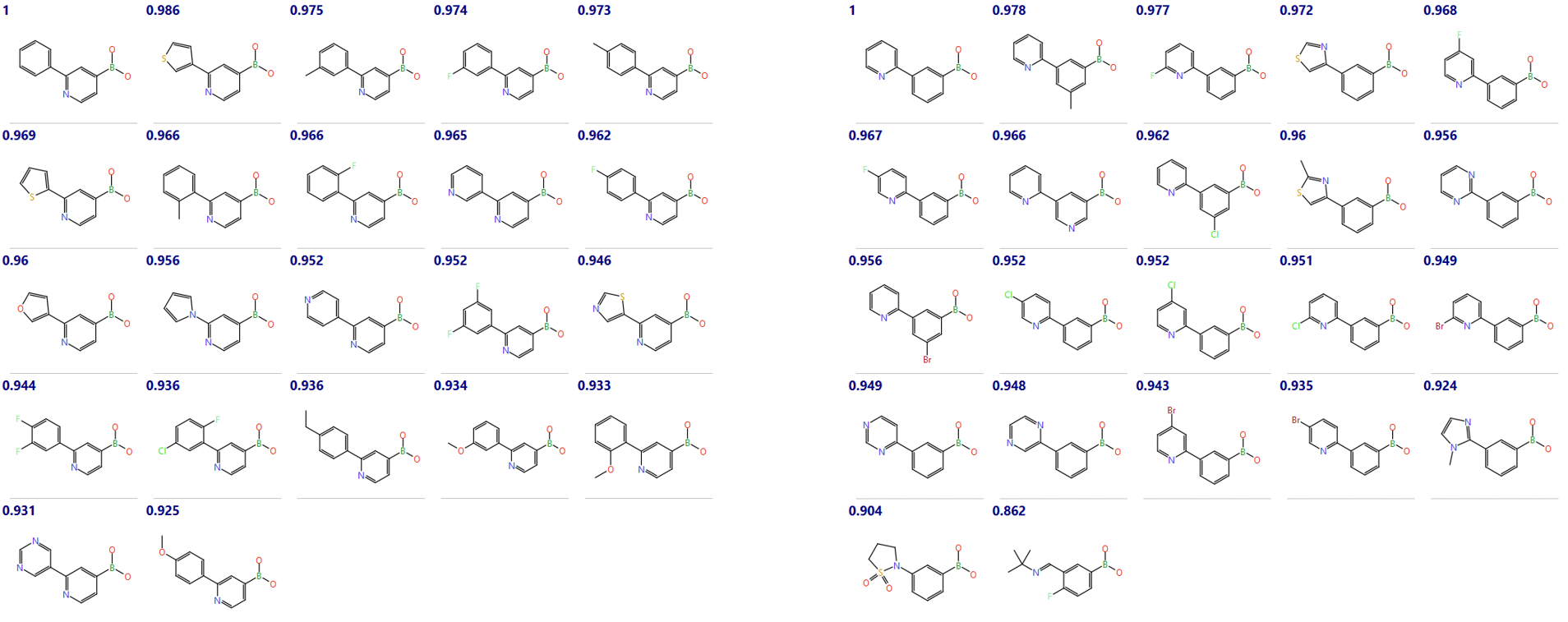
Figure 4: The utility of the method is neatly demonstrated by the populations of two of the top clusters which nicely separate 3-substituted phenyls from 3-substituted 4-pyridyls (shown below with similarity to medoid).
The full results are available on request from Cresset support.
PickR is an excellent method for clustering reagents for library design as it enables you to select electrostatically diverse monomers for high quality libraries and better intellectual property. The method considers conformational and electrostatic effects giving for a more diverse design of the reagent library. Although computationally more time consuming than traditional methods it can be applied to datasets of several thousand reagents easily using the built-in job distribution options.
Request an evaluation to try PickR on your project.