Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
On the tail of tropical storm Nestor, SERMACS, began in the historic colonial riverside city of Savannah Georgia (founded 1733). The meeting was attended by approximately 2,000 delegates from academic and research institutes in the southeastern United States.
There were several symposia focused on the areas of interest in the application of computational methods for drug discovery. This is a brief discussion of some of the presentations.
Robert Justin Grams, a graduate student of Dr David A. Brown, Chemistry, Virginia Tech, discussed development of peptidomimetic drugs based on the RYKF tetrapeptide, that stabilizes cardiolipin (an important major component of the inner mitochondrial membrane) for the restoration of mitochondrial structure-function and maintenance of inner membrane morphology. A designed mimetic Elamipretide1 (SS-31, MTP-131 and Bendavia) (D-Arg-dimethylTyr-Lys-Phe-NH2), currently being developed by Stealth BioTherapeutics, a cardiolipin peroxidase inhibitor and mitochondria-targeting peptide, has orphan disease treatment designation and is currently in Phase III trials for mitochondrial myopathy.
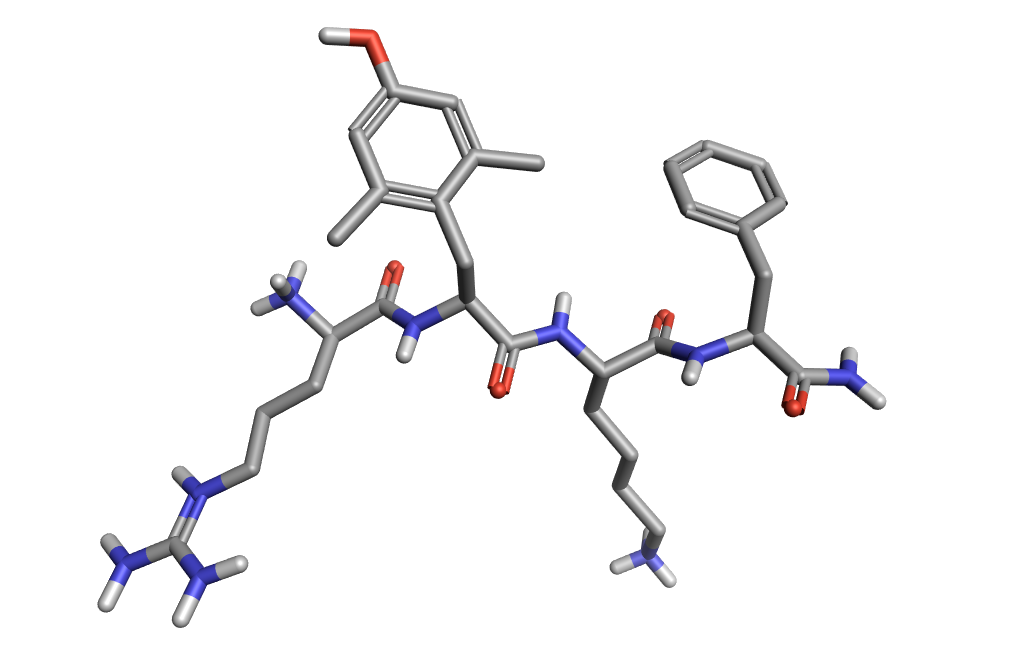 Figure 1: Elamipretide loaded into Forge™ and converted to 3D.
Figure 1: Elamipretide loaded into Forge™ and converted to 3D.
Dr Sadanandan Velu, University of Alabama, Birmingham, presented a drug target repurposing project. The development of VGSC subtype NaV1.5 subtype (NaV1.5) voltage-gated sodium channel blockers for breast-cancer metastasis inhibition2. VGSC channels in general, are already well-established targets for pain medications, however recently it has been noted that overexpression of NaV1.5 is associated with metastatic breast cancer (in MDA-MB-231 cells). Using Tetrodotoxin and known channel blockers, Dr Velu development a 3D-QSAR model (COMFA) and developed 5 lead compounds that had better activity as channel blockers and inhibitors of metastasis invasion than ranolazine and phenytoin. The most active compound blocked the sodium channel currents by 50% at 1uM.
Several undergraduate research groups, including one at High Point University in Winston-Salem NC, described impressive drug discovery research projects for undergraduate teaching institutions. Dr Meghan Blackledge described a drug repurposing project developing novel inhibitors of STK1, a ‘PASTA’ kinase (penicillin binding protein and Ser/Thr associated) which control genes involved in the development of antibiotic resistance. The group repurposed Loratadine and demonstrated that it could inhibit SKT1 and prevent beta lactam expression, and that inhibition of SKT1 prevents PBP2 (penicillin binding protein) expression in hospital strains.
The Structure-based Drug Design symposium was sponsored by Cresset. Dr Dev Arya, Clemson University, South Carolina, began by discussing the development of antimicrobial compounds targeting bacterial and fungal ribosomal RNA. Aminoglycosides bind to rRNA but have poor selectivity. Dr Arya and his group developed an rRNA targeted high throughput screening assay using aminoglycosides,3 like neomycin, bound to fluorescence probes. This assay provided a rapid screening method for a designed peptidic-aminoglycoside based chemical library targeting ribosomal RNA. They were able to identify antimicrobials against wild type and mutant strains that showed differences in binding affinity for the EColi A site and human A site RNA targets. Dr Arya also illustrated some modeling studies of compounds bound to the nucleic acids.
Dr Rajendra Tangallapally, St. Jude Children’s Hospital, discussed structure-based design of pantothenate kinase modulators for the treatment of CoA deficiencies. Panthothenate kinase is a metabolic enzyme which regulates CoA levels, and mutations in any of the 3 human PANK genes leads to PKAN, pantothenate kinase associated neurodegeneration. A screen of the St. Jude compound library followed by chemical modification lead to the development of PZ-2891 which is a CNS penetrant PANK allosteric activator that binds at the enzyme dimer interface (as shown in figure 2)4. Animal testing indicates drug administration increases CoA levels in mouse models. Dr Tangallapally indicated that there was no computational scaffold hopping tools used in the further design of molecules tested based on the PZ-2891 lead compound. Dr Tangallapally did discuss use of the Central Nervous System Multiparameter Optimization Desirability (CNS MPO Score) in his ligand design developed by Pfizer and based on ClogP, ClogD, TPSA, MW, HBD, pKa values.5
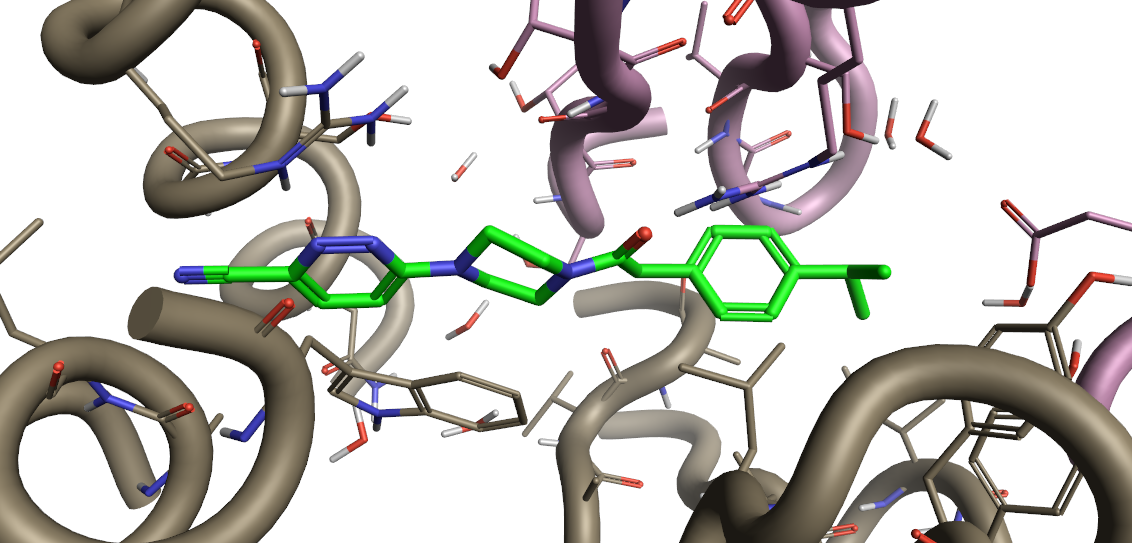
Figure 2. PZ-2891 from PDB 6B3V loaded in Flare.
Dr Campbell McInnes, Drug Discovery and Biomedical Sciences at The University of South Carolina, presented results on the development of type IV inhibitors of B-Raf kinases that block dimerization and overcome MEK/ERK activation. Using the dimerization interface of the Raf kinase, as a drug target, orally absorbed macrocyclic peptides based on a loop region present at the dimerization have been designed. High affinity B-Raf DIFs (dimerization interface) inhibitors can block signalizing in malignant cells. A fragment strategy called REPLACE with a fragment ligated inhibitor peptide has been applied to replace peptide elements with more drug like elements. Lead compounds that are type IV kinase selective inhibitors will be used to develop BRAF inhibitors applying these techniques to macrocyclic drug discovery.6
Dr Dmitriv Kireev, UNC Chapel Hill School of Pharmacy discussed the development of his FRASE database.7 FRAgments in Structural Environments (FRASE) which takes chemical information from structural and chemogenomic databases to create a kinase pocket design database for controlled polypharmacology to selectively target specific tyrosine kinases (TYRO3, AXL, MERTK(TAM). The full FRASE database used in this study was made available to public as a shared archive.
A FRASE includes information from a high affinity ligand and sequence alignment data. Several FRASEs extracted from different protein-ligand complexes are combined. A FRASE is a chemical fragment from a 3D protein bond ligand pose, if the fragment is good in one protein it will be good in another protein that has an identical structural environment. This can be used to design a new ligand for a kinase. It involves identifying the structure environment from the database that matches the binding pocket of the kinase of interest and then looking at the ligand fragments belonging to the FRASE and combining them into synthetic compounds. A case study was presented.
At this stage I presented our work on Electrostatic Complementarity™ highlighting it’s usefulness in small molecule design. For further detail, refer to our recent publication in J. Med. Chem. ‘Electrostatic Complementarity as a Fast and Effective Tool to Optimize Binding and Selectivity of Protein–Ligand Complexes’.8
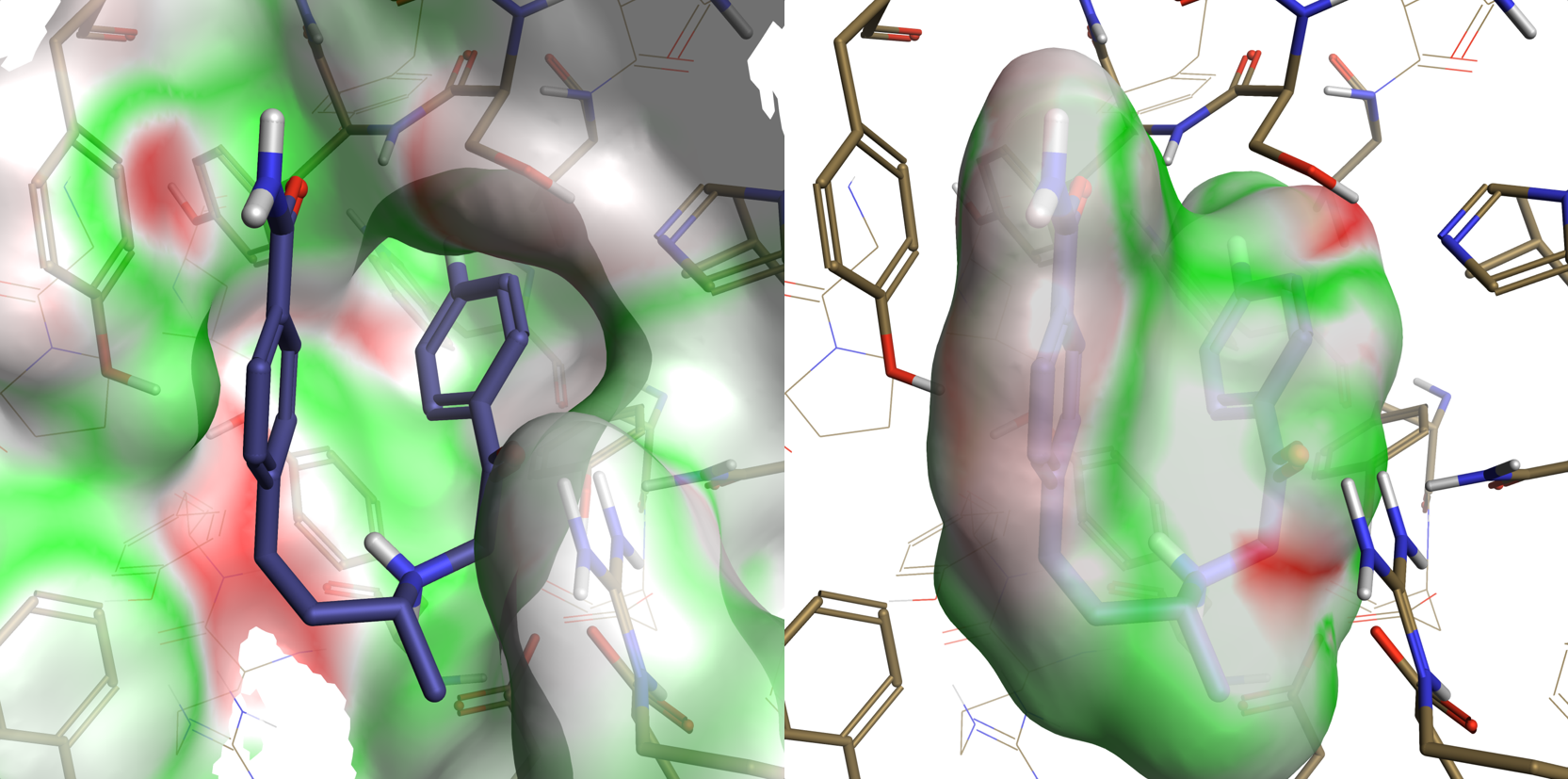
Figure 3. Electrostatic Complementarity of ligand in PDB 2OGZ as expressed on the protein surface (left) and the ligand surface (right).
Dr David Ostrov, University of Florida College of Medicine, discussed immunodulatory HLA binding small molecules.9 Dr Ostrov reported on the identification of the HLA-DQ8 as a drug target for diabetes. Employing UCSF Dock and a screen of the NCI Therapeutics Program compounds, several small molecules were identified as binders to HLA-DQ8 and triggering an HLA related immune response. These are under study for the prevention of type I diabetes which is an autoimmune disorder linked to HLA-DQ8.
Dr Pieter Burger, Emory University, reported on allosteric modulation of the NMDA receptor: identifying and exploring novel binding pockets. NMDA is a heterotetrameric complex. There are several PDB structures :4PE5, 4TLM and 4TLL although they are all poor resolution structure (3.59-3.96 angstroms resolution). Dr Burger developed an NMR program NAMFIS to perform NMR analysis of molecular flexibility in solution and given an ensemble of molecules the individual conformers could be extracted. ROC Scores were used with MD to improve docking. The Zinc database was used with Rocs followed by Glide docking, and MMGBSA to yield 12 compounds. A combination of modelling, and NMR-based methods were used to determine the bioactive binding pose of a series of compounds to an NMDA heteromeric structure.10
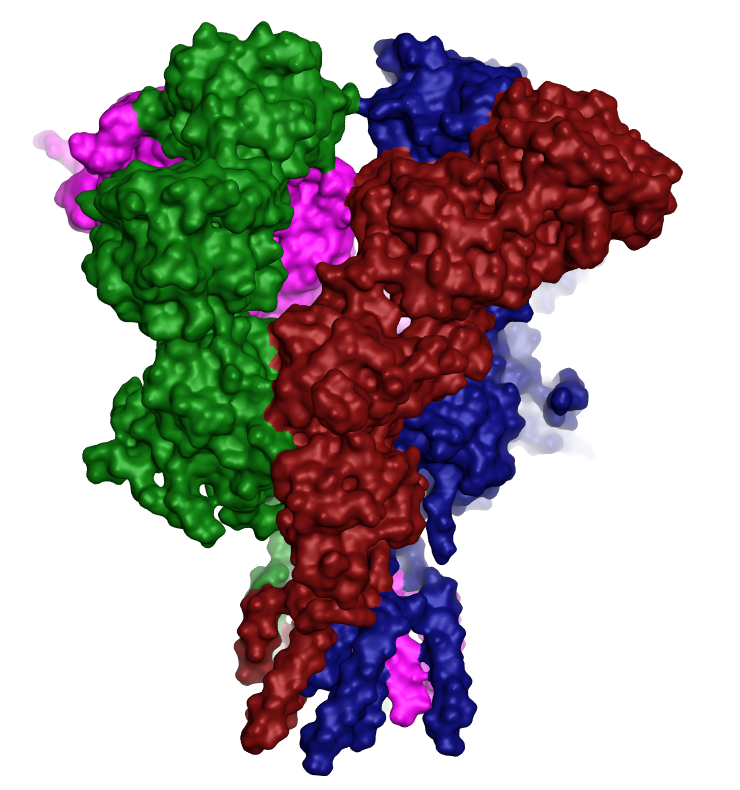 Figure 4. PDB 4PE5 loaded into Flare™ demonstrating the tetrameric nature of the NMDA receptor.
Figure 4. PDB 4PE5 loaded into Flare™ demonstrating the tetrameric nature of the NMDA receptor.
Dr Dennis Fourches, NC State, discussed how structure-based approaches are quickly evolving from classical 3D docking to multi-step, ensemble protocols involving a combination of flexible docking, machine learning and molecular dynamics. Dr Forches discussed an MD-Based Descriptor Matrix used to fit to a neural net MD QSAR model11. The main purpose of his studies was to understand the dynamics of protein-ligand interactions. The first example presented was the conformation of experimental GalU inhibitors for Listeria using virtual screening and molecular docking. The second example was a QSAR study using machine learning and chemical descriptors computed for every timestep of a molecular dynamics trajectory. This method was applied to a set of ERK2 kinase inhibitors and Bcr-ABl tyrosine kinase inhibitors. This is a novel concept to include dynamics in examining ligand selectivity in protein-ligand interactions. The third example was the use of molecular docking study of HLA-B*57:01 for the prediction of adverse drug reactions.
The computational drug design symposium was well attended and included many interesting questions and discussions.