Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
For a few years now the RDKit UGM has been of the great things about September, together with blackberries and Vuelta España, but I admit the latter two can be subjective.
This year's edition was kindly hosted by the University of Hamburg, and very well organized by Emanuel Ehmki and the other members of the local committee (Theresa Cavasin, Uschi Dolfus, Anna Lina Heinzke, Tim Kuhrt), plus of course Greg Landrum.
I won't spend many words on what the RDKit is; I doubt there are barely any computational chemists/cheminformaticians who have never typed a
from rdkit import Chem
command in a Python script or Jupyter Notebook of theirs. Indeed, Greg reported that a search in GitHub for the above Python line returns over 8500 code results across more than 450 unique repositories (i.e., excluding all RDKit forks), while Google Scholar counts over 550 hits for 'RDKit' in 2018 alone.
These numbers are impressive, and say a lot about how pervasive and essential the RDKit has become throughout the scientific community when it comes to deal with molecules on a computer.
Cresset was one of the gold sponsors of the UGM, as we believe the RDKit is a very important asset for our company and the whole cheminformatics community; we also contribute code when possible.
For instance, at this UGM I gave a lightning talk regarding the implementation of a new parameter in the Maximum Common Substructure (MCS) finding algorithm which enforces consideration of ring fusion.
I had observed that the RDKit returns the expected MCS between 1-methylnaphthalene and 2-aminonaphthalene, i.e. the naphthyl core (11 atoms, 10 bonds; Figure 1).
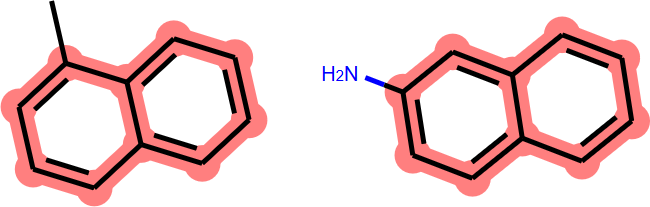
Figure 1. MCS between 1-methylnaphthalene and 2-aminonaphthalene (10 atoms, 10 bonds).
However, the MCS computed between 1-methylnaphthalene and 2-methylnaphthalene is the methylcyclodecapentaene envelope (Figure 2).
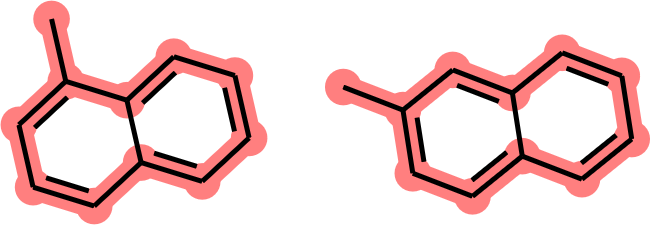
Figure 2. MCS between 1-methylnaphthalene and 2-methylnaphthalene (11 atoms, 10 bonds, no consideration of ring fusion).
While this is indeed the largest possible MCS between the two structures, it neglects the fusion bond between the two phenyl rings. As a chemist, I would have preferred to rather obtain the same naphthyl core as before (Figure 3).
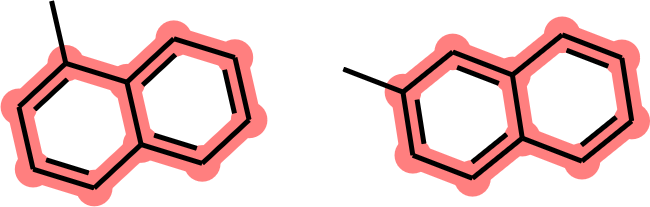
Figure 3. MCS between 1-methylnaphthalene and 2-methylnaphthalene (10 atoms, 10 bonds, takes ring fusion into account).
So I added a new parameter to the MCS algorithm to enable this behavior that I believe is more useful and intuitive from a chemist's perspective. My code is currently under review and, once approved, it will be merged into the main codebase.
But – this was supposed to be a blog post about the RDKit UGM, not about me ranting on MCS! So let's move on to the main highlights from the talks without further ado. As usual, this review is far from comprehensive and mostly focuses on the talks which came with code attached.
Greg gave the usual overview of the new features introduced in the latest release, as well as some sneak peeks into features which are likely to be incorporated in the forthcoming release.
Among others, I really loved this nifty Jupyter Notebook integration gem, i.e. the possibility to interactively select atoms by clicking on a 2D sketch, and to then retrieve the indices of those atoms (Figure 4). Greg had already anticipated this in a post on the RDKit blog, and I believe it can really help to bring one step further the interaction with 2D molecules in a notebook.
Figure 4. An example of how atoms can be selected in a Jupyter Notebook and used to rotate a dihedral identified by those atoms.
I also liked the fact that a format to store/retrieve atomic properties to/from a SDF file was introduced (Figure 5). Anyone dealing with partial charges, atom names and the like has had to do something along these lines, but having a standard way of doing it within the toolkit is of course much better.
The upcoming integration with NextMove's MolHash functionality is very promising to compare molecules independently of their tautomeric or resonance form – looking forward to seeing it implemented in the next release of RDKit.
Finally, the fact that the RDKit-powered SMILES/SMARTS/InChI key searches and substructure searches are now available through Google Patents is simply fantastic – Ian Wetherbee (Google) had already presented this at the San Diego ACS meeting, and also gave a lightning talk here to illustrate the new functionality.
Figure 6. Google Patents now supports chemistry-aware searches through the RDKit.
Dominique Sydow and Jaime Rodríguez-Guerra presented some sterling work done in Andrea Volkamer's group in Berlin around providing a series of talktorials built on top of Jupyter Notebooks and KNIME workflows to teach CADD topics using open-source software. The main concept about these teaching resources is that they break the barrier between theoretical lectures and hands-on, practical sessions, as you learn doing things at the same time you actually do them. Jupyter Notebooks allow interleaving explanatory text with code (Figure 7), and similar considerations also apply to the KNIME-based workflows even though through a different paradigm.
I can imagine how much displaying rich output close to the input helps keeping students engaged during interactive coding sessions: I wish I had such tools available when I was in academia! The next generation of data scientists and computational chemists is truly in good hands.
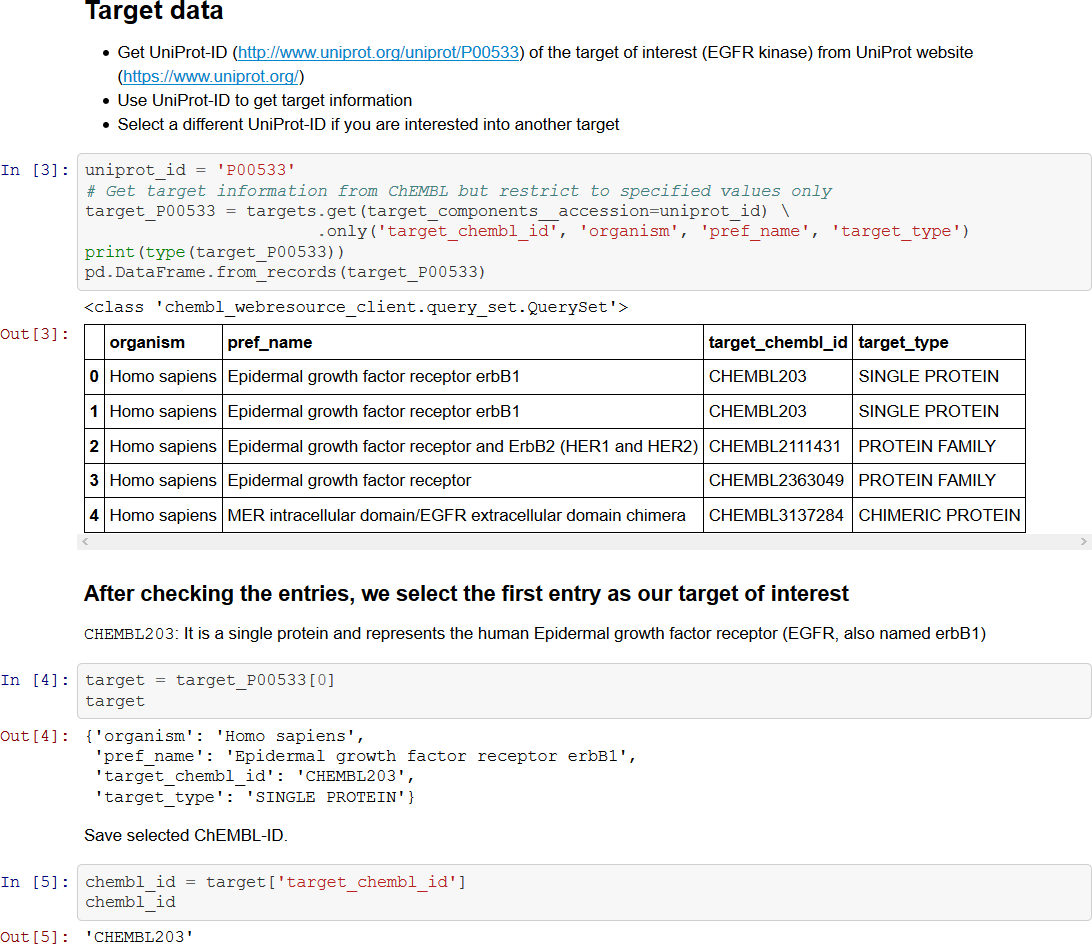
Figure 7. An excerpt from a Jupyter Notebook teaching how to extract data from ChEMBL using its Python API and display it using Pandas dataframes.
Among other things, Rob Smith (Heptares) presented a ChemSketch-to-RDKit molecule converter that he implemented internally to facilitate conversion of 2D molecule sketches made by medicinal chemists into ready-to-dock 3D structures. The interesting bit is that the initial implementation, before ACD provided a PDF document describing the file format, was made by reverse engineering the file format using a hex editor, which is always an entertaining exercise I have done myself earlier in my career. Rob is going to contribute the sk2 parser to the RDKit as soon as he gets internal approval.
Jan Halborg Jensen (University of Copenhagen) presented some applications of the RDKit to quantum chemistry. He started by presenting a couple of earlier works on prediction of pKa values and prediction of the regioselectivity of electrophilic aromatic substitution reactions, where the RDKit was used to enumerate protonated isomers via reaction SMARTS, conformational searches and to display results. Jan also presented an xyz-to-RDKit molecule converter, able to turn a table of atomic elements with Cartesian coordinates and a total formal charge (i.e., the typical input and output of a QM program) into an RDKit molecule with correct bonds, bond orders and localized formal charges (Figure 8). I have tested xyz2mol in Flare™ starting from an xyz representation of zwitterionic bilastine and it indeed did a perfect job at placing double bonds and formal charges in the correct places:
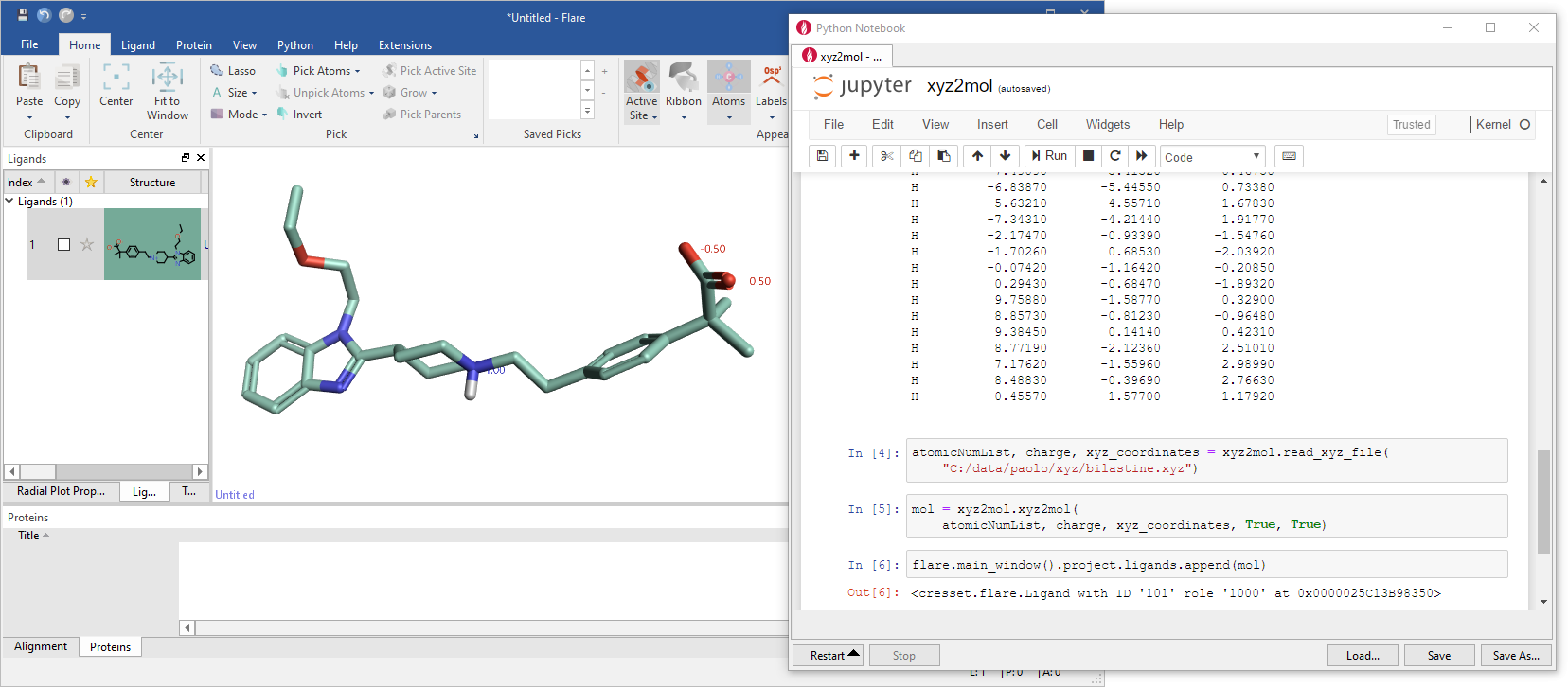
Figure 8. Jan Jensen's xyz2mol in action in Flare.
While this has not yet been thoroughly tested on a large dataset, it has already proven very useful when used on internal research projects. Finally, he introduced a graph-based genetic algorithm and generative model/Monte Carlo tree search for the exploration of chemical space, where insertion/deletion of atoms, changes in bond order and other molecular graph editing operations are all handled via RDKit-driven reaction SMARTS.
Also Christoph Bauer (University of Bergen) applied the RDKit to QM calculations. His work focused on generating bimolecular hydrogen-bonded complexes by enforcing proper distance and angle restraints and then doing a constrained MMFF minimization with the RDKit (Figure 9).
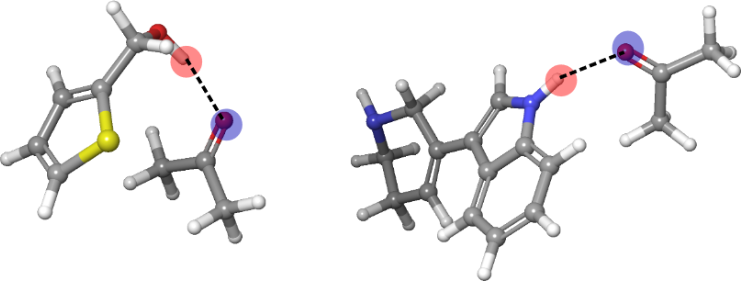
Figure 9. Structures of hydrogen-bonded complexes generated and optimized using the MMFF implementation provided by the RDKit.
These complexes were then subjected to QM calculations and used to train a machine learning model to predict hydrogen bond strengths.
The main idea which sparked the grow3D project, part of Torx™, was: how nice would it be to draw a molecule in your favorite 2D sketcher and simply see it grow sensibly within the binding site in the 3D viewport, naturally obeying to the constraints imposed by the active site, with little or no intervention from the user? The RDKit is extensively used throughout the grow3d workflow for substructure matching, MCS finding, file parsing, etc. Read the poster.