Background
RNA is undruggable - right? Medicinal chemists have almost entirely focused on protein targets to treat human disease and we have developed numerous strategies to discover small molecules that interact with enzymes, receptors and ion channels. However, these only form a tiny proportion (1-2%) of the landscape in the post-genomic era. RNA, the product of 70-80% of genome transcription, is now attracting attention as a drug target. Of course, many RNA biologics – aptamers, siRNA, antisense oligomers and synthetic mRNA, have been in clinical development for years, but despite major investment in this area only two examples have gained clinical approval. Both treat rare diseases: nusinersen (2016) for spinal muscular atrophy (SMA), the most common genetic cause of paediatric mortality, is an antisense oligomer; and patisiran (2018) the first RNAi drug treats hereditary transthyretin amyloidosis, a rare liver disease. In fact, the discovery that conjugation of oligonucleotide drugs to the amino sugar GalNac enhances uptake into hepatocytes makes liver diseases an obvious target for RNA biologics, but progress in developing drugs for other diseases has been less successful.
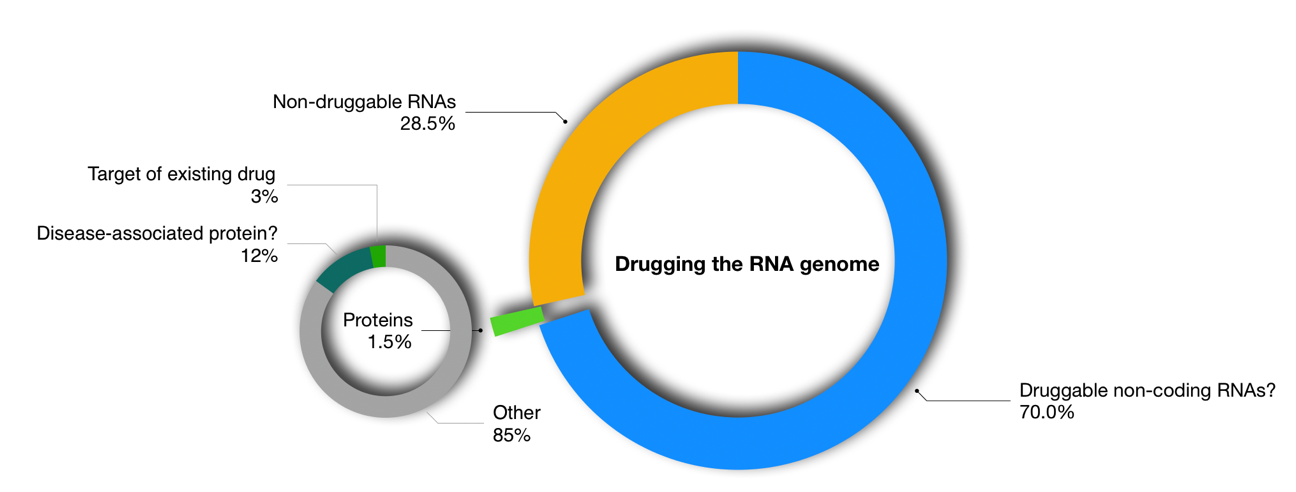 Figure 1: Only 1.5% of the human genome encodes proteins. Of these 10-15% are considered to be disease associated and current drugs target ~3% of them i.e., 0.05% of the human genome. Some 70% of non-coding RNAs represent potential drug targets so targeting these could increase the scope for small molecule drug discovery.
Figure 1: Only 1.5% of the human genome encodes proteins. Of these 10-15% are considered to be disease associated and current drugs target ~3% of them i.e., 0.05% of the human genome. Some 70% of non-coding RNAs represent potential drug targets so targeting these could increase the scope for small molecule drug discovery.
So, is there a role for small molecules in this field? As with protein, RNA forms complex 3D-structures and has its own repertoire of structural features such as hairpins, pseudoknots, internal loops and bulges which form where there are perturbations of the A-form helix. Indeed, the Protein Databank contains > 4,500 examples of RNA structures, just under 3% of the current total, many of which contain co-crystallized small molecules located in binding pockets. So it is clearly possible for a small molecule to bind to RNA. Antibiotics are a case in point where around half of FDA approved drugs target the ribosome or riboswitches, so we have had drugs that target bacterial RNA for decades. That said, our ability to design drugs that target human RNA targets is still in its early phase.
Examples of RNA targeting small molecules
One good example is Branaplam from Novartis, an orally active pyridazine (mwt 393) in Phase 2 trials for SMA (July 2019), the same target as nusinersen. It acts by modifying the splicing pattern of the SMN2 gene, which compensates for loss of function in the SMN1 gene, the hallmark of the disease. The overall effect is to increase levels of survival of motor neuron protein and prevent neurodegeneration. Initial hits were identified by screening the Novartis compound collection (1.4 M compounds) albeit with <1% hit rate.1 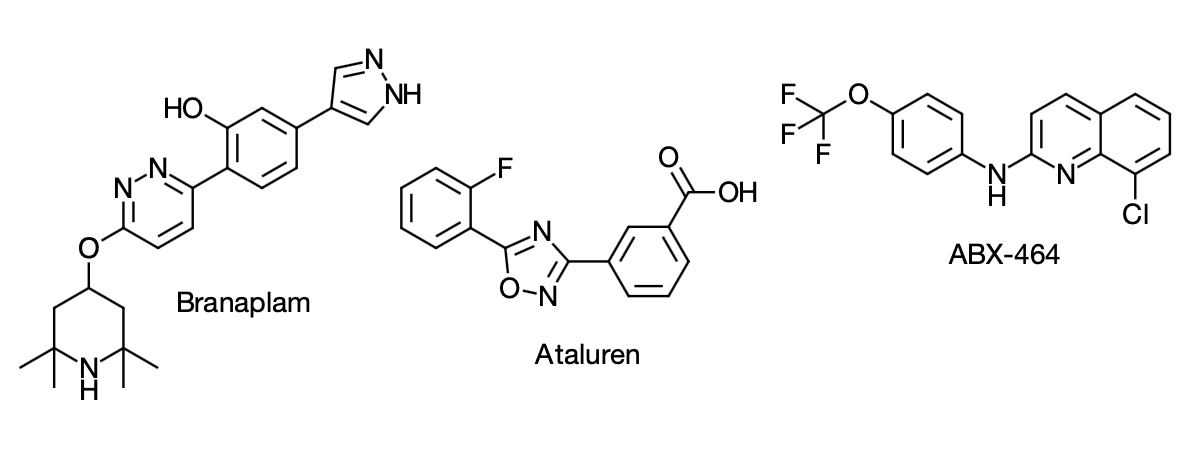 Figure 2: Small molecules in clinical development for human RNA targets.
Figure 2: Small molecules in clinical development for human RNA targets. 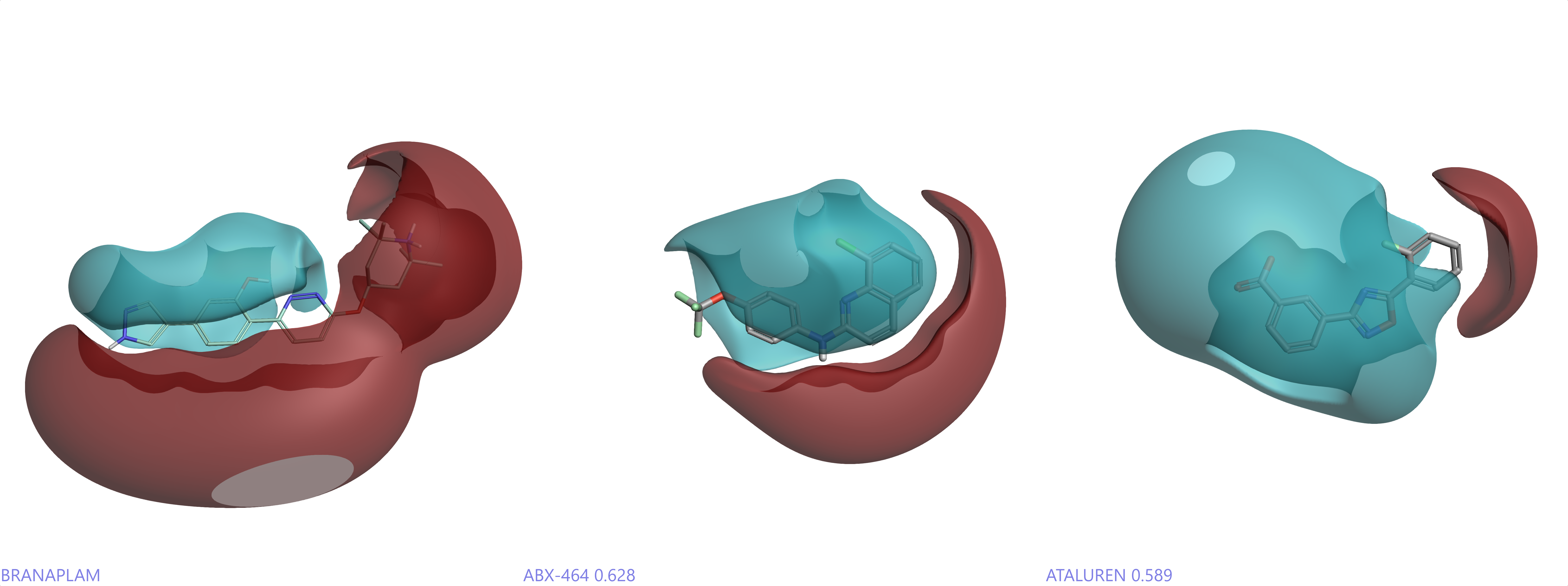
Figure 3: Cresset derived positive (red) and negative (cyan) electrostatic fields of Branaplam1 (left), Ataluren3 (middle) and ABX-46422 (right).
A second recent example is the discovery of ABX-464 from Abivax which is being evaluated for treatment of ulcerative colitis.2 The compound binds to the cap binding complex and acts by increasing expression of miR-124, a microRNA precursor, which decreases inflammation. Preliminary results of a Phase 2a trial in a small group of 32 patients were not statistically significant, but 30% of the treated group were in clinical remission (as were 11% of the placebo group). Nevertheless, Abivax hope that this discovery paves the way to a safe and efficient treatment in a range of inflammatory diseases.
The most advanced compound is Ataluren,3 from PTC therapeutics, which was licensed to treat Duchenne’s muscular dystrophy in Europe in 2014 but has not yet been approved by the US FDA. Full details of the oxadiazole’s mechanism of action are not known but it effectively increases levels of dystrophin by bypassing the stop codon in people with a nonsense mutation in the dystrophin gene.
Matt Disney’s group at Scripps in Florida have identified compounds that bind to the site recognized by the ribonuclease Drosha, an essential enzyme for converting pri-miRNAs to pre-miRNAs. They identified a molecule that binds sites on both miR-515 and miR-885 with similar affinity. Noticing that an adjacent binding site exists in miR-515 they linked two molecules together and obtained a dimer (TGP-515) that binds selectively to 515 with an affinity of ~60 n.4 The final molecule is not .’small.’ by medicinal chemistry standards but was still effective when applied to cells in vitro. miR-515 represses generation of sphingosine kinase 1 and the group were able to demonstrate that TGP-515 increased levels of both kinase and sphingosine-1-phosphate, important in many processes, including inflammation, cell proliferation and migration. The upshot was to increase levels of Her2, which is regulated by the SK1/S1P pathway but not normally expressed in MCF-7 breast cancer cells, making them sensitive to treatment with Herceptin. Normal breast epithelial cells do not express miR-515 and so this represents a highly targeted approach to treating breast cancer.
Novel approaches
In particular, Disney’s group have focused on establishing methods for predicting RNA binding sites for small molecules, described in his recent ACS Perspective5. These include 2-Dimensional Combinatorial Screening (2DCS) a protocol for screening an immobilized array of small molecules against a labelled library of RNA motifs. Their strategy has identified a number of approved anti-cancer drugs, principally kinase and topoisomerase inhibitors, that bind to RNA in addition to the protein targets for which they were originally developed. This has been coupled with a method for assessing fitness scores i.e., a measure of the affinity and selectivity of the interaction of the small molecule with the RNA motif (StARTS), to give a platform named Inforna whose aim is to predict potential compounds based on RNA sequence alone.
It is notably easier to predict RNA secondary structure from primary sequence by free energy minimisation than for proteins. Algorithms such as ViennaFold 2.466 (last updated in 2017) provides rapid predictions at around 75% accuracy, considerably better than the success rate for proteins.
A particular strength of the Cresset XED force field7,8 is its ability to model p-p stacking interactions and indeed some of our earliest studies applied it to DNA structure. Given that RNA and DNA molecules, large and small, are governed by polar and stacking interactions, electrostatics (Figure 3) are key considerations for small molecules that will interact with them. In addition, Cresset’s structure-based modeling solution, Flare™, and techniques therein such as Electrostatic Complementarity™, are highly applicable to these systems.
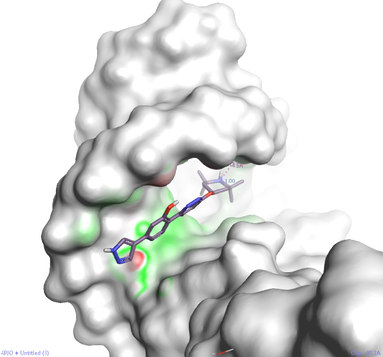
Figure 4: Cresset’s Electrostatic Complementarity surface (green good / red bad - electrostatic complementarity score) for a Branaplam binding pose, in the RNA groove of human U1SNRNP derived from PDB: 4PJO. It was previously thought that, unlike DNA, the major and minor grooves of RNA were not optimal binding sites for small molecules, but the Novartis group presented compelling biophysical data suggesting that Branaplam binds to the major groove of dsRNA.
Strategies and tools developed by these groups are fundamental to our understanding of how to drug RNA. It is becoming clear that medicinal chemists have much to contribute to this area and that our knowledge of the pharmacokinetics and dynamics of small molecules is equally applicable to the RNA universe beyond the confines of the protein world.
Request a free confidential discussion
To see how Cresset Discovery Services can add value to your project, contact us for a free confidential discussion.
References
- It was previously thought that, unlike DNA, the major and minor grooves of RNA were not optimal binding sites for small molecules, but the Novartis group presented compelling biophysical data suggesting that Branaplam binds to the major groove of dsRNA. Palacino, J. et al. sMn2 splice modulators enhance U1–pre-mRNA association and rescue SMA mice Nat. Chem. Biol. 2015, 11, 511–517
- Vermeire, S et al.,ABX464 is safe and efficacious in a proof-of-concept study in ulcerative colitis patients, European Chron’s and Colitis Organisation conference 2019 abstract OP21
- Welch, EM et al., PTC124 targets genetic disorders caused by nonsense mutations, 2007, Nature, 87-91.
- Costales, MG et al, A Designed Small Molecule Inhibitor of a Non-Coding RNA Sensitizes HER2 Negative Cancers to Herceptin J. Am. Chem. Soc. 2019 141 7 2960-2974.
- Disney MD, Targeting RNA with small molecules to capture opportunities at the intersection of chemistry, biology, and medicine. J. Am. Chem. Soc. 2019 141 (17), 6776-6790 and references therein.
- Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011, 6, 26.
- Molecular Field Extrema as Descriptors of Biological Activity: Definition and Validation. J. Chem. Inf. Model. 2006, 46 (2), 665-676.
- https://www.cresset-group.com/science/overview/#xedff.
 Figure 1: Only 1.5% of the human genome encodes proteins. Of these 10-15% are considered to be disease associated and current drugs target ~3% of them i.e., 0.05% of the human genome. Some 70% of non-coding RNAs represent potential drug targets so targeting these could increase the scope for small molecule drug discovery.
Figure 1: Only 1.5% of the human genome encodes proteins. Of these 10-15% are considered to be disease associated and current drugs target ~3% of them i.e., 0.05% of the human genome. Some 70% of non-coding RNAs represent potential drug targets so targeting these could increase the scope for small molecule drug discovery. Figure 2: Small molecules in clinical development for human RNA targets.
Figure 2: Small molecules in clinical development for human RNA targets. 
