The emergence of targeted protein degradation in drug discovery
In recent years, targeted protein degradation has emerged as a new therapeutic modality. This new technology is now part of many research and development programs for pharmaceutical and biotech companies, with programs now reaching pre-clinical and clinical development. It provides a new approach for drug targets that were previously thought to be undruggable such as transcription factors, scaffold proteins, or proteins that lack or have a broad or shallow binding pocket.
Cresset Discovery has experience in targeted protein degradation, and has supported companies in this space globally. Our team of CADD experts can provide a deep insight into your degrader system by applying specialist computational approaches and expertise.
What are protein degraders?
Targeted protein degradation broadly refers to strategies that harness the cell’s own machinery to eliminate or degrade a protein of interest, instead of inhibiting it. There are a number of different strategies currently being used to develop protein degraders.
Proteolysis targeting chimeras (PROTACs) and molecular glues are two major approaches to promote proteasome-targeted protein degradation, the cellular waste disposal system 1,2. Both methods operate by bringing the targeted protein in close proximity to an E3 ubiquitin ligase enzyme, such as Cereblon (CRBN) or von Hippel-Lindau protein (VHL), or to any of the 600 known E3 ubiquitin ligases, to engage the ubiquitin-proteasome system (UPS) by forming a ternary complex (shown in Figure 1).
The targeted protein is then ubiquitylated by the ligase protein, marking it for proteasomal recognition and degradation. After, the degrader molecule dissociates and is recycled to target a new copy of the targeted protein. This catalytic-type mechanism of action has some advantages over traditional small molecule inhibitors, as most degraders such as PROTACs require a lower concentration to effect the same response which reduces the incidence of off-target and potentially toxic effects.
![]()
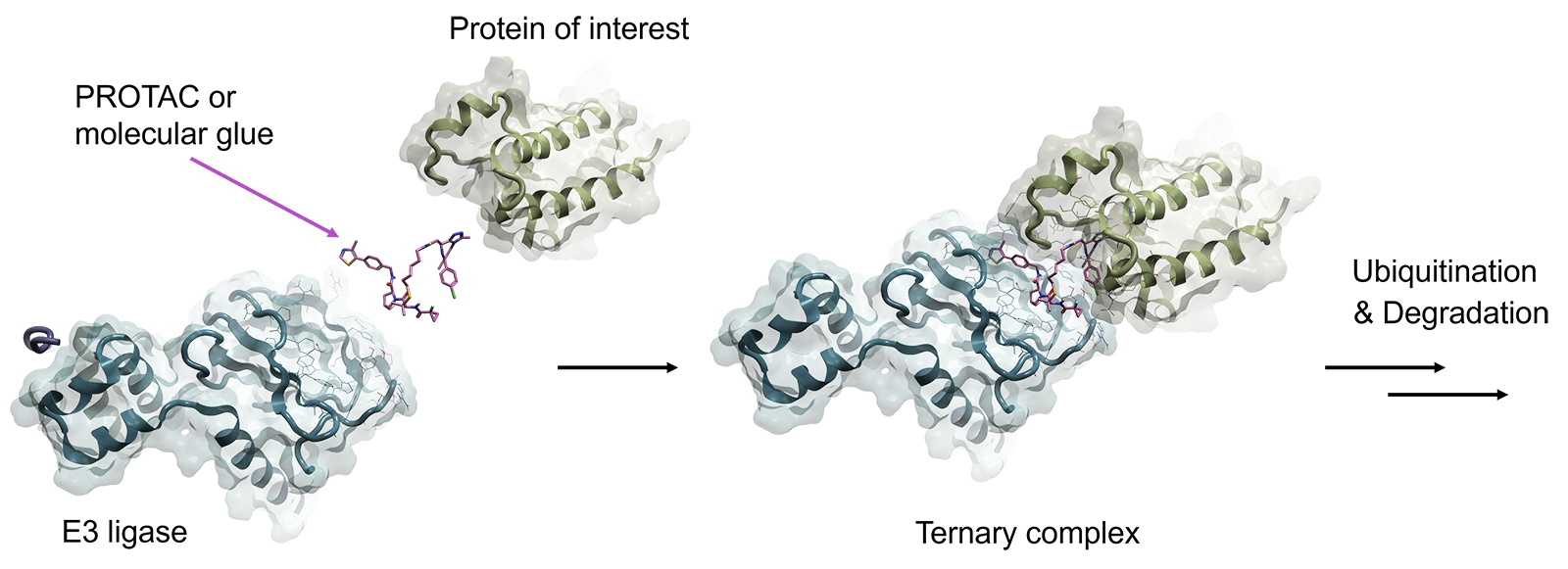
Figure 1. A PROTAC molecule (pink) binds in a ternary complex with its protein target (green) and the E3 ligase (blue) to trigger proteasomal degradation
PROTAC molecules are heterobifunctional small molecules consisting of two ligands connected with a flexible or rigid linker, with one ligand binding to the targeted protein and the other ligand binding to E3 ubiquitin ligase. Whereas molecular glue degraders stabilize the interactions between E3 ubiquitin ligases and the protein of interest by engaging PPI (protein-protein interaction) directly between the two proteins.
Optimize protein degrader design with computational chemistry expertise
You can enhance every step of the design and development of your degrader by engaging support from our expert modelers. Not only are we experienced in identifying selective degraders of disease-causing proteins as well as new E3 ligase warheads, but we will undertake diligent structure-guided design of your degrader molecules, including evaluation of its ternary complex and the impact of the linker. We can help with the choice of the E3 ubiquitin ligases and consider the physicochemical properties of degraders that may go beyond Lipinski’s ‘rule of five’ scope.
Many experimental structures of ternary complexes are available for computational investigations on degrader binding 2. Our team utilise these structures to help you to visualize and understand PPIs as well as interactions with the degrader molecule and offer invaluable insights to optimize the design of new degraders. Where an experimental structure is unavailable, that of a suitably related protein or proteins may be employed to construct an overlay model or a homology model.
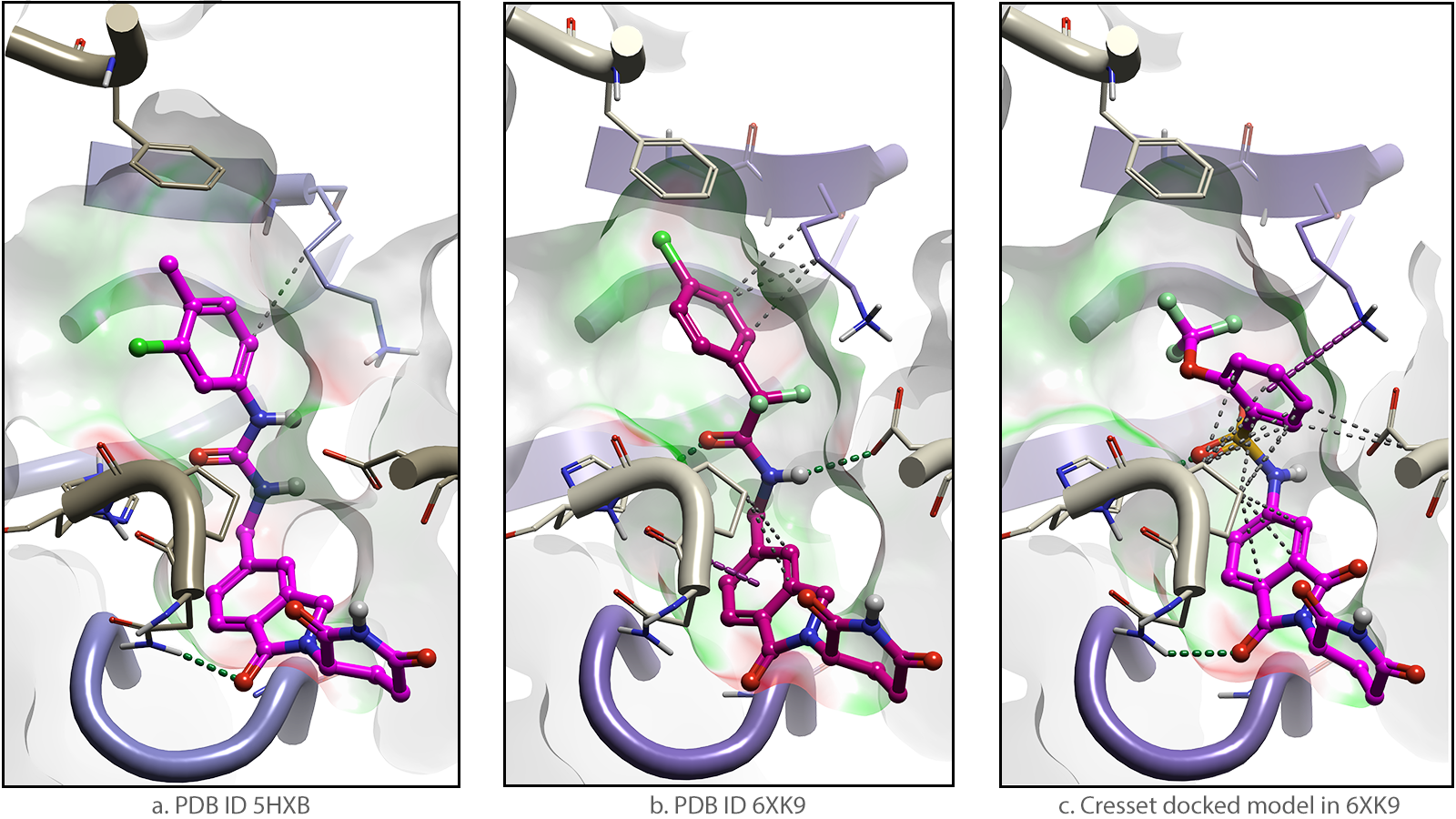
One example is the design of glue degraders that complex GSPT1 to CRBN. Phenotypic screening and library design identified the early GSPT1 degrader CC-885 (Figure 2a) and subsequently CC-90009, also known as Eragidomide that is currently in Phase 1 trials for acute myeloid leukemia (Figure 2b) 3. A separate group who then applied molecular docking and structure-based library design reported the improved degrader, SJ6986 (Figure 2c) 4.
![]()
![]()
Figure 2. Molecular glue degraders that complex CRBN (beige) to GSPT1 (purple)![]()
![]()
![]()
![]() By modeling the interactions attained by the GSPT1 degrader ligands 5, we can provide visualization of the molecular surfaces of the experimental and docked complexes. The regions colour-coded in green are where the electrostatic field of the degrader complements that of CRBN. These areas become more extensive as GSPT1 degraders are progressively optimized. Correspondingly, the red regions with unfavourable EC are reduced.
By modeling the interactions attained by the GSPT1 degrader ligands 5, we can provide visualization of the molecular surfaces of the experimental and docked complexes. The regions colour-coded in green are where the electrostatic field of the degrader complements that of CRBN. These areas become more extensive as GSPT1 degraders are progressively optimized. Correspondingly, the red regions with unfavourable EC are reduced.
Cresset scientists incorporate EC predictivity in many protein degradation modeling workflows, using proprietary methods. Where the structure of the protein receptor is available, we score EC alongside the orthogonal docking score to assess accurately and rapidly thousands of hits 6. In library design, we rank enumerated structures to shortlist ideas for visual inspection and identify those with favorable interactions with the receptor or similarity to a known ligand. In virtual screening, we have been identifying novel active ligands with notable success 7. Our established design paradigms are equally applicable to the E3 ubiquitin ligase and the target receptor for degradation. This also extends to modeling chemical replacements for a specific degrader component such as the ligase warhead or a PROTAC linker.
Engage Cresset Discovery to accelerate your targeted protein degradation project
Cresset Discovery serves as a true partner for computational modeling in drug discovery. Our expert team has a breadth of experience across medicinal and computational chemistry, and biology to work alongside you to deliver your project goals.
To learn more about how we can support you in a protein degradation project or any other project, contact us or request a no-risk free confidential discussion with our team.
References
- Miklos Bekes, David R. Langley and Craig M. Crews, PROTAC targeted protein degraders: the past is prologue, Nature Reviews Drug Discovery 2022, 21, 181-200. https://www.nature.com/articles/s41573-021-00371-6.
- Zuzanna Kozicka and Nicolas Holger Thoma, Haven’t got a glue: Protein surface variation for the design of molecular glue degraders, Cell Chemical Biology 28, 1032-1046. https://doi.org/10.1016/j.chembiol.2021.04.009.
- Surka, C., Jin, L., Mbong, N., Lu, C.-C., Jang, I.S., Rychak, E., Mendy, D., Clayton, T., Tindall, E.A., Hsu, C., et al. CC-90009, a novel cereblon E3 ligase modulator targets acute myeloid leukemia blasts and leukemia stem cells, Blood 2020,137, 661–677. https://doi.org/10.1182/blood.2020008676.
- Nishiguchi, G., Keramatnia, F., Min, J., Chang, Y., Jonchere, B., Das, S., Actis, M., Price, J., Chepyala, D., Young, B., McGowan, K., Slavish, P.J., Mayasundari, A., Jarusiewicz, J.A., Yang, L., Li, Y., Fu, X., Garrett, S.H., Papizan, J.B., Kodali, K., Peng, J., Pruett Miller, S.M., Roussel, M.F., Mullighan, C., Fischer, M., Rankovic, Z., Identification of potent, selective, and orally bioavailable small-molecule GSPT1/2 degraders from a focused library of cereblon modulators J. Med. Chem. 2021, 64, 7296–7311. https://doi.org/10.1021/acs.jmedchem.0c01313.
- Electrostatic Complementarity™ (EC)
Flare™, Cresset®, Litlington, Cambridgeshire, UK; https://www.cresset-group.com/flare/; Cheeseright T., Mackey M., Rose S., Vinter, A.; Molecular Field Extrema as Descriptors of Biological Activity: Definition and Validation J. Chem. Inf. Model. 2006, 46 (2), 665-676; Bauer M. R., Mackey M. D.; Electrostatic Complementarity as a Fast and Effective Tool to Optimize Binding and Selectivity of Protein–Ligand Complexes J. Med. Chem. 2019, 62, 6, 3036-3050; Maximilian Kuhn, Stuart Firth-Clark, Paolo Tosco, Antonia S. J. S. Mey, Mark Mackey and Julien Michel Assessment of Binding Affinity via Alchemical Free-Energy Calculations J. Chem. Inf. Model. 2020, 60, 6, 3120–3130
- Lead Finder™, BioMolTech®, Toronto, Ontario, Canada; https://www.cresset-group.com/lead-finder/
- Blaze™, Cresset®, Litlington, Cambridgeshire, UK; https://www.cresset-group.com/blaze/; Cheeseright, T.J.; Mackey, M.D., Melville, J.L.; Vinter, J.G. FieldScreen: Virtual Screening Using Molecular Fields. Application to the DUD Data Set. J. Chem. Inf. Model. 2008, 48 (11), 2108-2117; Cheeseright T., Mackey M., Rose S., Vinter, A.; Molecular Field Extrema as Descriptors of Biological Activity: Definition and Validation. J. Chem. Inf. Model. 2006, 46 (2), 665-676

