Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
The role of water is crucial in protein-ligand binding process. Ligands are often optimized towards replacing energetically unfavorable water molecules in the protein binding pocket in structure-based drug design. On the other hand, enthalpically favorable water molecules can be regarded as mediators between the protein and the ligand1, and protein-water-ligand interactions could also be important in the binding process. Therefore, understanding water stability in binding sites is crucial for successful structure-based drug design.
It is possible to determine the behaviour of water molecules in protein-ligand binding sites computationally. One of the water analysis methods implemented in Flare™ is Grid Inhomogeneous Solvent Theory (GIST)2. A restrained molecular dynamics simulation of the protein system is performed with explicit water, and the distribution and thermodynamics of water molecules of a predefined volume (usually the binding site) are calculated with inhomogeneous solvation theory (IST).
The first order entropies and first- and second-order energies can be obtained from the IST theory. The solvation entropy is approximated by the solute-water correlation as shown in Equation (1), where kB is the Boltzmann's constant, ρo is the number density of bulk solvent and gsw(r,w) is the solute-water pair-correlation function.
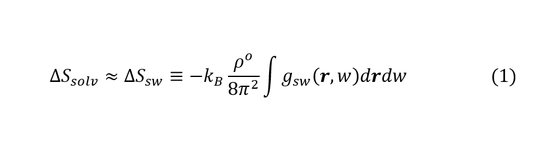
The solvation energy ΔH is obtained via the water-water energies with respect to bulk water and solute water energy calculated with the MD force field. The solvation free energy can be computed following Equation (2):
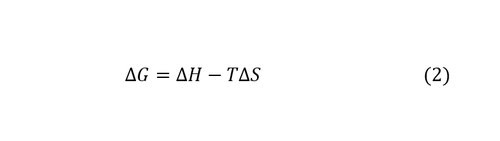
Three-dimensional mappings of water density and the above thermodynamic properties are generated from GIST calculations. Regions of high and low water stability can be highlighted from the three-dimensional grid maps, which can be used as a tool to reveal hydration sites and to guide computer-aided ligand design (Figure 1).
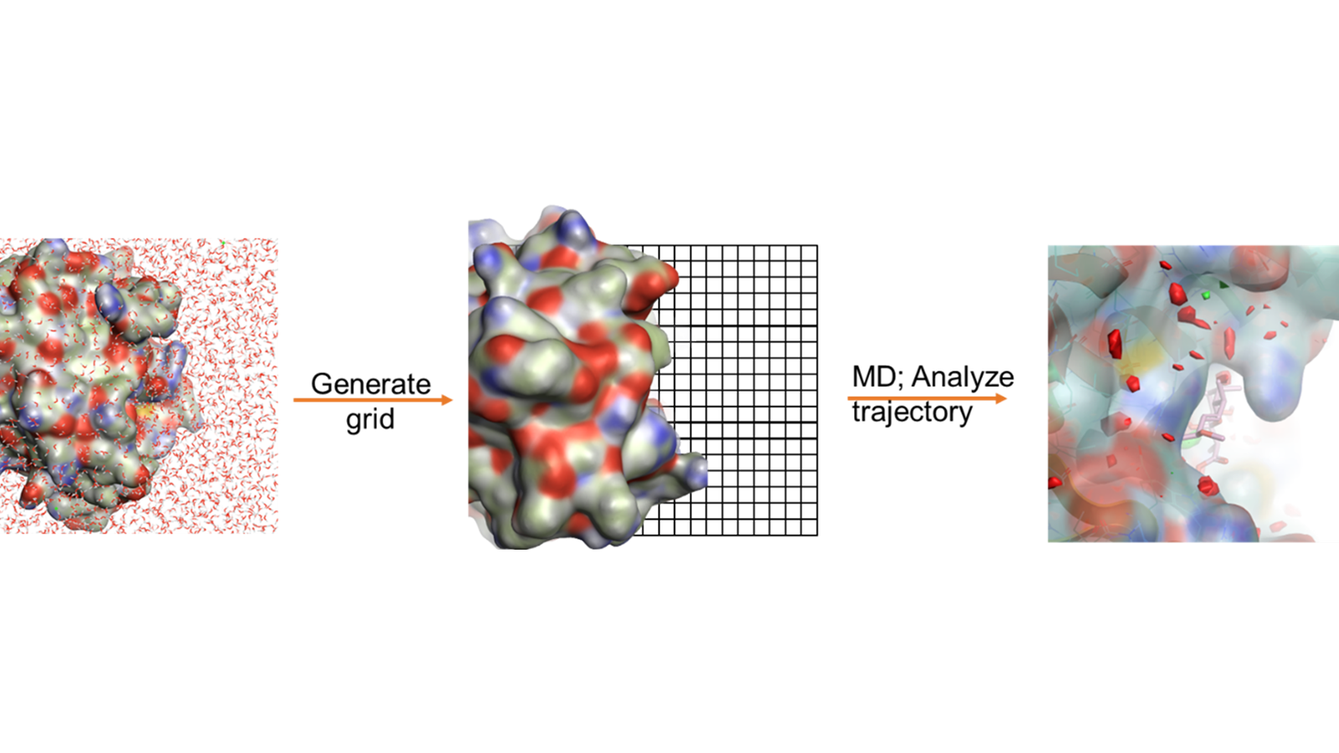
Figure 1: Workflow of GIST and the heat surface map of water stability obtained from GIST calculation.
Application of GIST in protein-ligand binding and ligand design is still being explored. The additional water analysis features for GIST calculations in Flare V6 provide guidance on multiple perspectives, such as desolvation, energetic and entropic contributions.
GIST water analysis experiments on HSP-90 (PDB: 3RLP) are shown in Figure 2 (apo structure, ligand shown for reference only) and Figure 3 (holo structure with ligand 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine). It can be seen from Figure 2a that water molecules 4 and 9490 are both ‘unhappy’ with positive solvation free energies, indicating that those water molecules could be replaced or displaced by fragments in the ligand. However, both water molecules are enthalpically ‘happy’, indicating strong interaction with the protein or surrounding water molecules. For the purpose of ligand design, polar functional groups correctly positioned with similar enthalpic interaction with the environment could replace such water molecules.
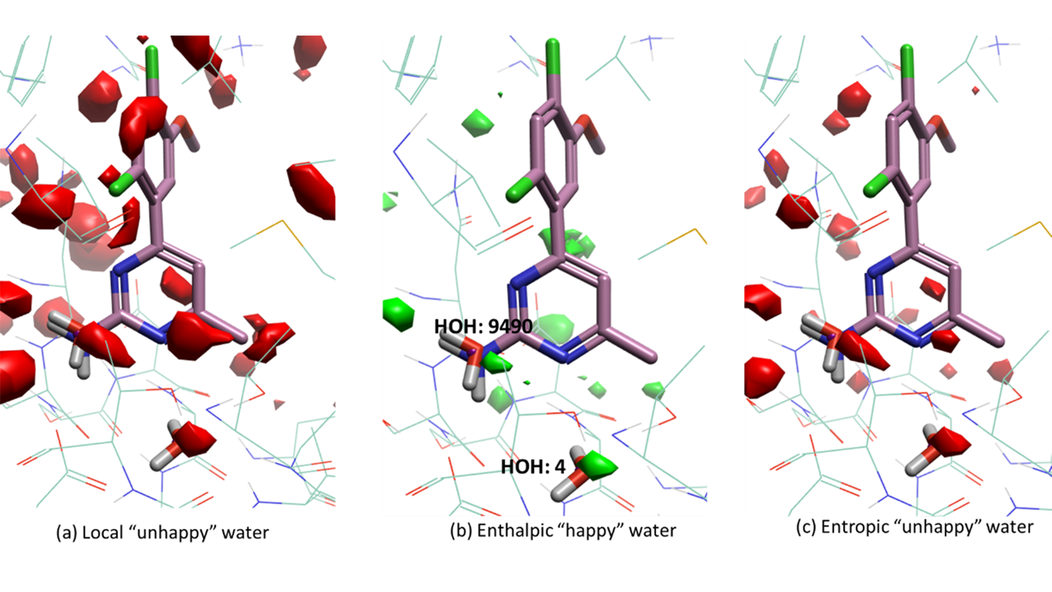
Figure 2: GIST calculation result for apo structure of HSP-90 (PDB 3RLP), ligand structure shown for reference only. (a) Isosurface of local ‘unhappy’ with water density >= 4, dG >= 0.5 kcal/mol. (b) Isosurface of enthalpic ‘happy’ water with dH <= -2 kcal/mol. (c) Isosurface of entropic ‘unhappy’ water with -TdS >= 2 kcal/mol.
According to the GIST calculation on the protein-ligand complex (Figure 3), water molecule 9490 is replaced by the amino group of the ligand, and the amino group could make enthalpic interaction with the environment. However, water molecule 4 is located close to the heatmap of local ‘happy’ water in the holo structure. It can be seen from Figures 3b and 3c that water molecule 4 is enthalpic ‘happy’, and the enthalpic contribution outweighs the entropic penalty, which indicates that water molecule 4 is a bridging water molecule and is stabilized in the binding process.
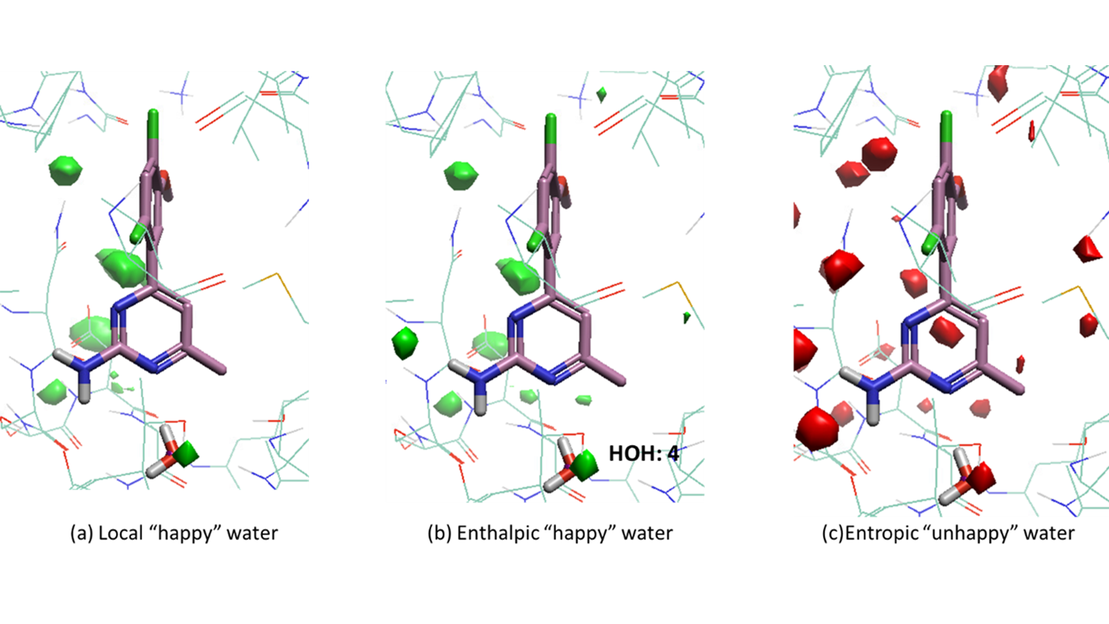
Figure 3: GIST calculation results for holo structure of HSP-90 (PDB 3RLP) with ligand 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine). (a) Isosurface of local ‘happy’ with water density >= 4, dG <= -0.5 kcal/mol. (b) Isosurface of enthalpic ‘happy’ water with dH <= -2 kcal/mol. (c) Isosurface of entropic ‘unhappy’ water with -TdS >= 2 kcal/mol.
An interaction map of the ligand with the surrounding environment is shown in 2D in Figure 4. Hydrogen bonds are formed between the ligand and residue ASP-A93 and water molecule 1 (replacing that for water molecule 9490 in Figure 1). Weak hydrogen bonds are formed between the ligand and water molecule 4, which stabilizes the water molecule as shown in Figure 3a.
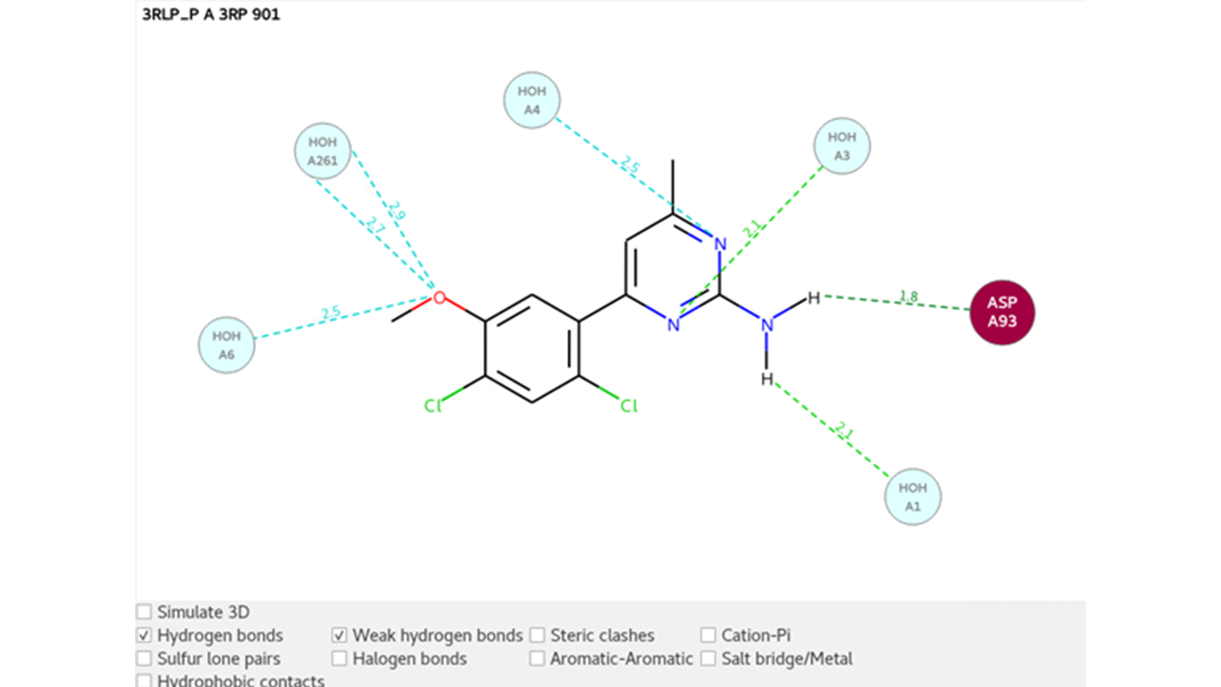
Figure 4: 2D interaction map of the ligand with surrounding environment with hydron bonds highlighted for PDB 3RLP.
GIST calculations are simple to set up and run. The improved display of GIST results in Flare V6 make interpretation of binding site waters significantly easier. It enhances the ability of the computational chemist to understand the properties of water in the active site and the effect of ligand binding on the stability of these waters.
If you would like to try GIST on your project, request an evaluation today.
Flare is a single platform for ligand-based and structure-based drug design that enables research chemists to discover novel small molecules efficiently and effectively. New features in Flare V6 include library enumeration, quantum mechanics and pocket detection analysis. Read the full release announcement.