Discovery
Deducing ligand-protein interactions
Deducing ligand-protein interactions is a piece of molecular detective work and the difficulty of the task depends on the starting point. In some cases, a protein structure will be available; in others there might be a molecule, a chemotype or a piece of protein or DNA. The task is to work out how they fit together. Of course, if a crystal structure is available then the job is almost done. A clear crystal structure shows the ligand’s active conformation in the protein binding site. When this is not available, modeling can help to fill in the missing pieces.
Clues to the protein active site
Any information about the protein, in conjunction with ligand data, can often tell you something about the active site. For example, you may have some mutagenesis data about what the protein does, or you may be using a chimera protein construct. Any data of this sort provides information about the functional active site – or indeed any site of potential use for small molecule intervention.
In addition, SAR data about the ligand can be very helpful for deducing the important features for interactions, whether these are the electrostatics, hydrophobics or H bonds. If you can work out what is important to the interaction, you can use it to piece together the data from the protein structure.
Arriving at an active ligand conformation hypothesis
Once the shape of the binding pocket has been determined, it has to be related to the shape and electrostatics of the active ligand conformation hypothesis.
Cresset Discovery uses the unique XED force field for analysing the shape, electrostatics and hydrophobicity of both the ligand and the protein. It’s a universal force field, so it doesn’t need parameterizing for different situations. This makes it ideal for ligand-protein minimization in order to check an interaction hypothesis.
The goal of ligand-protein minimization is to check which ligand conformation hypotheses are realistic. The goal is to arrive at a hypothesis that has low protein energy and low ligand energy before minimization, and then has a lower system energy when the protein and ligand are brought together for the minimization.
This method throws out any poor hypotheses. For example, a minimization may fail if it can’t reduce the total energy of the system. This can be the result of a residue being perturbed from a position where it has relatively low energy, in which case you know that you’ve got it wrong.
Case study: Medicine for Malaria Venture
One of our customers, Medicine for Malaria Venture (MMV), had found a ligand through a high throughput screen. They developed a compound and improved its activity, but then asked Cresset Discovery to work backwards from the compound to determine the probable site of action at the protein target. The antimalarial target has been published in this paper: Nature 522 pg 315-320 2015.
MMV already had some resistance mutagenesis data. They observed residue changes in the protein that caused it to resist the medication and were able to assign a few residues that were important for the recognition. This gave us some clues to work on.
Cresset Discovery took this starting data and used it to build a homology protein model of the malarial parasitic protein. We then looked at the structure-activity relationships around the molecules of interest, carried out conformation sampling of the molecule and looked at the residues and the pocket that was the likely place of interaction of the molecule. Using this we developed a binding hypothesis for the molecule.
There were nine different possibilities for the conformation of the molecule. We slightly adjusted each of the conformations, looked at some different permutations of how the residues were placed and how they were orientated in the pocket. Then we took each of the ligand-protein interaction hypotheses, minimized them and looked at the ligand-protein’s binding energies. Based on these, we could judge which of the hypotheses were reasonable.
On the basis of one of these hypotheses we carried out a virtual screening exercise and a fragment replacement exercise. The ultimate aim of the project was to provide MMV with another chemotype for them to work on so that they have a back-up series for their published series, preserving the novel mode of action they had discovered. MMV’s next step will be to buy the compounds or have them synthesised and get them tested. The work from Cresset Discovery gave them some good ideas and certainly gave them the best chance of finding something from commercial compounds that they can buy.
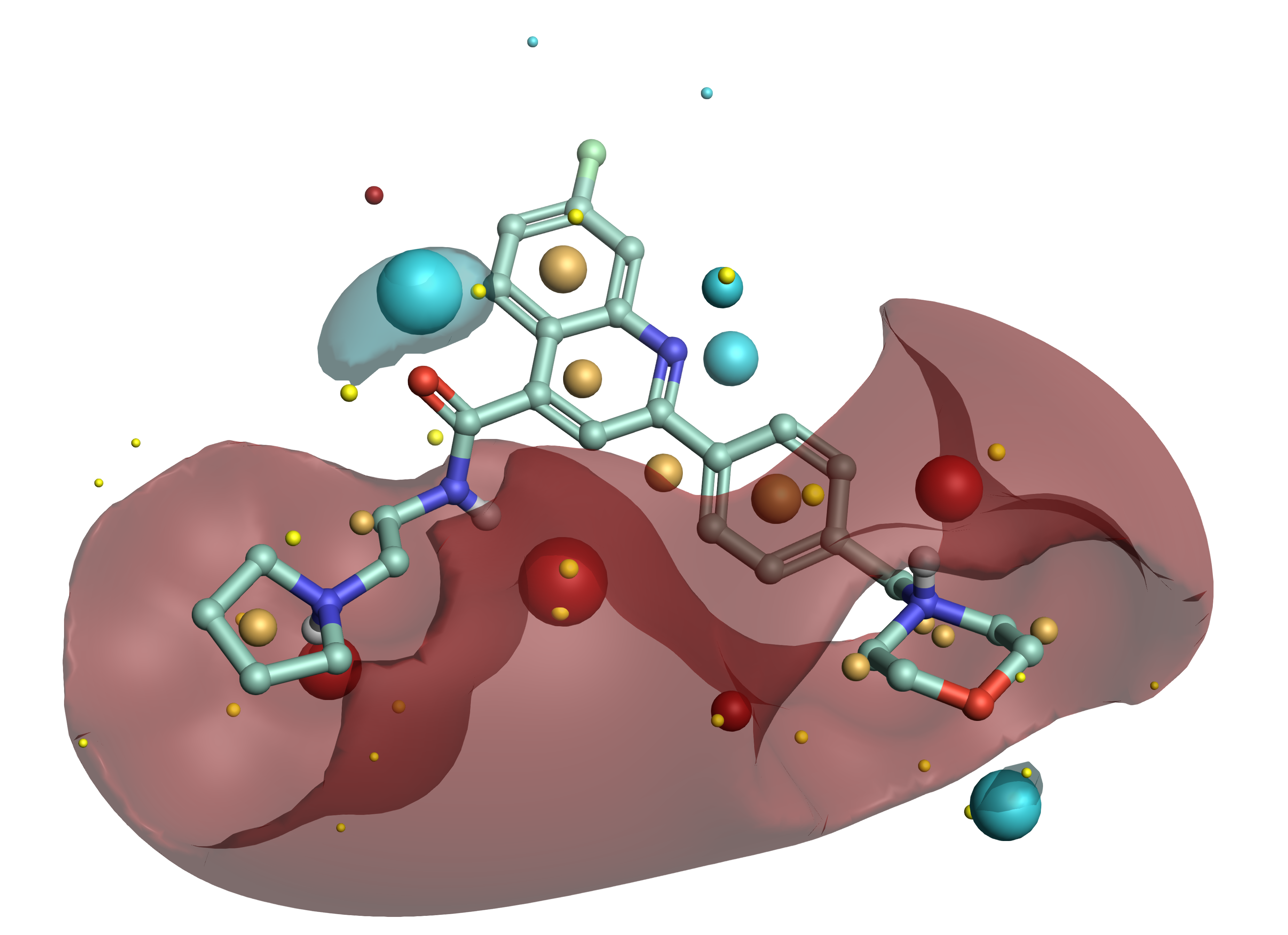
Example ligand from Medicines for Malaria Venture’s Nature paper showing a low energy conformation and Cresset field points generated using Forge™
Discovery
Contact us for a free confidential discussion
We help you reach your next milestone faster and more cost effectively
Contact us for a free confidential discussion