Discovery
Grow and link fragments
Cresset Discovery uses Spark for bioisosteric replacement. We believe that it is the best available tool for scaffold hopping and its bioisosteric replacement using fragment and reagent-specific databases can be readily applied to the popular discovery approaches of fragment growing and linking.
The starting point is one or more fragments that are known to be responsible for
F-uracil as an anti-cancer and anti-viral treatment
Uracil DNA glycosylase (UDG) is a potentially interesting target for both cancer and anti-viral therapies. Recent efforts to produce synthetic inhibitors of this protein relied on an active fragment tethering approach, yielding interesting bis-oxime linked active ligands. This example describes an alternative method using Cresset’s scaffold hopping software Spark for the reconnection of distant fragments.
To mimic a fragment linking experiment with the software, the bis-oxime linker was excised from an active ligand (PDB: 3FCI, Figure 2). The two fragment atoms to be joined were selected and suitable fragments with appropriate distance and geometry, which were capable of re-joining them, were inserted. Fragments were sourced from databases derived from ZINC and ChEMBL. The resulting molecules were automatically constructed in-situ, minimized and then scored against the parent compound using a field-based 3D similarity metric.
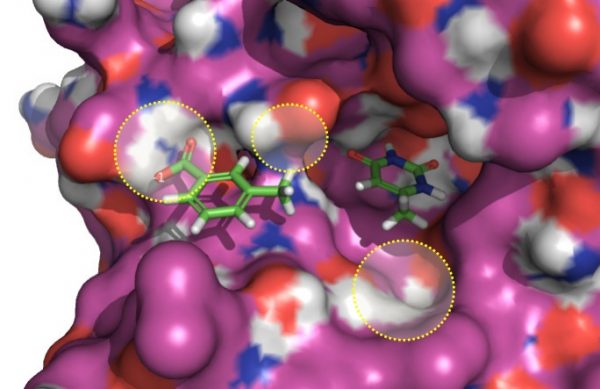
Active fragments originally tethered by a bis-oxime linker (disconnected for this experiment), shown embedded in UDG. In the
The example results (Figure 3) show the diverse range of the output suggestions for new linking chemistry. Importantly, each of the new fragments not only satisfies the geometry and length requirements for the reconnection of the
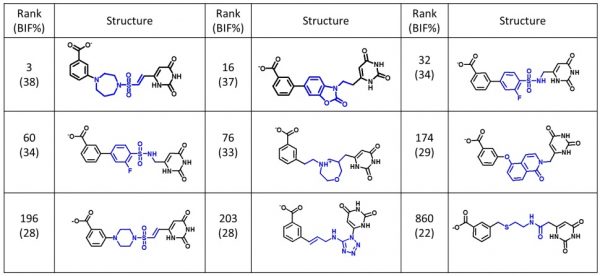
Example outputs from a fragment linking experiment using scaffold hopping in Cresset’s Spark.
Discovery
Contact us for a free confidential discussion
We help you reach your next milestone faster and more cost effectively
Contact us for a free confidential discussion