Discovery
Identifying allosteric pockets
Allosteric pockets can have a significant effect on the mechanism of protein action. Identifying and understanding them is particularly important for targets where traditional drug design methods fail and when HTS screens reveal unexpected results.
When does an understanding of allosteric pockets become important?
Many active sites for some very interesting drug targets are undruggable using traditional active site drug design methods. This is usually due to problems of selectivity, pocket architecture or the number of different biological pathways the protein is involved with. For example, in the blog Alzheimer’s disease: Modeling current and potential targets we describe how the secretase pathway is not exploitable as a potential Alzheimer’s drug target by traditional methods due to off-target effects. When the protein active site is not amenable to drug discovery, identifying allosteric pockets can present a different method to perturb the protein function and rescue the project.
Allosteric pocket identification can also be extremely helpful in another specific case. It can happen that hits resulting from a high-throughput screening (HTS) campaign are incompatible with the known active site. The identification of allosteric pockets and their effect on the mechanism of action is critical for any future development of such a lead and the understanding of protein function. 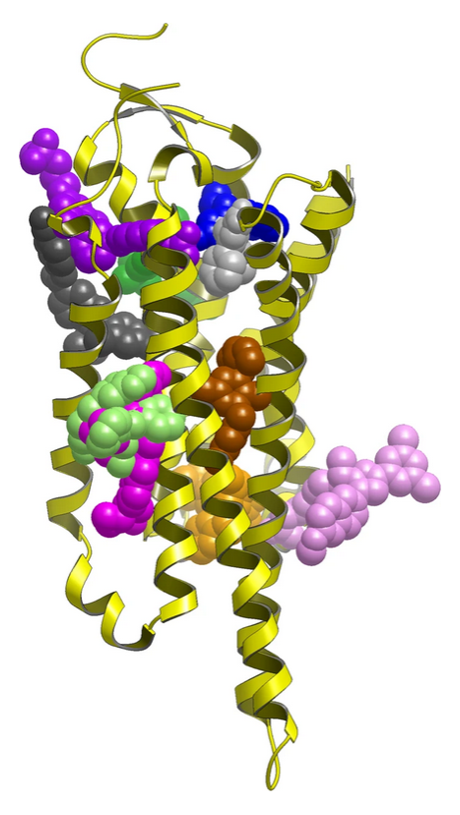
Figure 1: Experimentally validated allosteric sites in GPCRs1.
Allosteric pockets and protein function
Allosteric pockets are distinct from the protein active site but disrupt the protein function of interest by a non-traditional active site occupancy mechanism. The effect on the target protein of an allosteric modulator can be via any of the following:
- stabilizing an inactive conformation
- interfering with protein-protein binding
- blocking co-factor binding
- prevention of conformational rearrangements required for function.
Allosteric pockets have the advantage of tending to be unique to the protein of interest so selectivity tends to be far less of a problem, which can be critical when tackling proteins with common co-factors like ATP, or phosphate binding active sites in general. Allosteric modulators have lots of advantages, but the identification of suitable allosteric pockets is difficult to predict and often not obvious from the crystal structure data. They tend to be transient and dependent on the crystallographic conditions, so they are not always obvious from a single static crystal structure. ![Phosphate binding site (right) and allosteric binding site (left) for Tyrosine-protein phosphatase non-receptor type 11 (SHP2), displayed using Flare™[2] for the 6mdb[3] and 4pvg[4] crystal structures. Phosphate binding site and allosteric binding site for Tyrosine-protein phosphatase non-receptor type 11](/media/uploads/files/Phosphate_binding_site_and_allosteric_binding_site_for_Tyrosine-protein_phosphatase_non-receptor_type_11.png)
Figure 2: Phosphate binding site (right) and allosteric binding site (left) for Tyrosine-protein phosphatase non-receptor type 11 (SHP2), displayed using Flare™2 for the 6mdb3 and 4pvg4 crystal structures.
There are multiple software packages that will identify pockets on a protein surface and rank and score them for drug amenability. However, for a pocket to be useful for allosteric drug discovery it must also influence the protein function in which we are interested, and this is beyond the scope of these packages. Casual manual examination of most if not all proteins in the PDB database will reveal multiple pockets on the protein surface which are druggable, however, this does not make them suitable as an allosteric pocket as most will not perturb the protein behaviour of interest. It is only by combining an understanding of protein function, movement and interaction analysis that we can hope to identify those pockets which are allosteric to the biological process of interest.
Protein investigation and the hunt for allosteric clues
The first step in the identification of an allosteric pocket is, therefore, to step back and to fully understand the protein function, movements and interactions with co-factors, ligands and other proteins that are important for the protein mechanism of interest. Examination of other members of the protein family can yield valuable information on protein function and also on regions of the protein which are non-conserved and highlight the regions which have the potential for mechanistic selectivity. Only by having a full understanding of the protein behaviour and its interactions can we successfully identify allosteric pockets which are of relevance to the biological pathway of interest under consideration.
![Example protein movements upon ATPbinding for the biotin carboxylase subunit of pyruvate carboxylase using crystal structures 1ulz[5] for the apo from (left) and 1dv2[6] for the APT bound form (middle). The right hand image shows the two proteins aligned by the lower domain only to highlight the general tertiary structural conservation and the key ATP recognition conformational changes displayed in Flare. Example protein movements upon ATPbinding for the biotin carboxylase subunit of pyruvate carboxylase using crystal structures](/media/uploads/files/Example_protein_movements_upon_APT_binding_for_the_biotin_carboxylase_subunit_of_pyruvate_carboxylase.png)
Figure 3: Example protein movements upon ATPbinding for the biotin carboxylase subunit of pyruvate carboxylase using crystal structures 1ulz5 for the apo from (left) and 1dv26 for the APT bound form (middle). The right hand image shows the two proteins aligned by the lower domain only to highlight the general tertiary structural conservation and the key ATP recognition conformational changes displayed in Flare2.
Ideally there will be multiple protein crystal structures available displaying multiple states of the protein from which to build mechanistic hypotheses. Models of protein movement or protein-protein interactions greatly aid the identification of allosteric sites, highlighting key regions of the protein. However, even in the case where only one form of the protein is known, if there is sufficient understanding of the protein function and mechanism then hypotheses can be built to identify allosteric pockets.
It may also be possible to build a homology model of an alternate form of the protein if suitable templates are available to generate the required protein form. Molecular dynamics can also assist in this process by investigation of protein conformational flexibility and allowing the investigation of protein ligand stability upon binding. Pockets which are obviously undruggable can be ignored even if they are allosteric as there will be no opportunity for lead development.
Assessing opportunities
Once we have identified a set of potential allosteric pockets, increased focused evaluation of these sites for druggability can be undertaken using a combination of software and our own experience. This allow us to evaluate the pocket dispassionately via the software and also to apply knowledge and experience of tackling drug targets to critically evaluate the pockets and select those which present the best opportunity for progressing the project forward. The strength of evidence can be used to prioritize the order of analysis, but all identified pockets should be analysed to provide the maximum understanding and value.
Outcome
At the end of this process a set of potential allosteric pockets should be identified, plus the mechanistic justification for the selection of the pockets. The identified potential allosteric pockets can then be ranked using the gathered evidence before undergoing further analysis. The result of this process should be a set of potentially allosteric pockets with mechanistic evidence for their mode of action, and an estimate of the druggability of the pockets from which to enable a final informed decision about which pockets to pursue.
Where next?
How to proceed will depend on the starting position. If the starting point was an HTS hit then an analogue can be suggested to generate SAR and validate the allosteric site hypothesis. Alternatively, if there is no chemical matter, a virtual screening campaign can be carried out to provide a set of suggestions to hopefully provide the starting chemical matter from which to build SAR and allosteric pocket validation.
If you have a difficult target that could benefit from this type of computational analysis, or you have an HTS hit which is inconsistent with your active site, contact Cresset Discovery for a free consultation to find out how we can advance your project.
References
- Wakefield, A., Mason, J.S., Vajda, S. et al. Analysis of tractable allosteric sites in G protein-coupled receptors. Sci Rep 9, 6180 (2019) doi:10.1038/s41598-019-42618-8
- Flare structure-based design platform https://www.cresset-group.com/software/flare/
- 6-Amino-3-methylpyrimidinones as Potent, Selective, and Orally Efficacious SHP2 Inhibitors. Sarver, P., Acker, M., Bagdanoff, J.T., Chen, Z., Chen, Y.N., Chan, H., Firestone, B., Fodor, M., Fortanet, J., Hao, H., Hentemann, M., Kato, M., Koenig, R., LaBonte, L.R., Liu, G., Liu, S., Liu, C., McNeill, E., Mohseni, M., Sendzik, M., Stams, T., Spence, S., Tamez, V., Tichkule, R., Towler, C., Wang, H., Wang, P., Williams, S.L., Yu, B., LaMarche, M.J. (2019) J Med Chem 62: 1793-1802
- Crystal structure of protein tyrosine Shp2 catalytic domain complex with compound L88N79 Zhang, Z.-Y., Zeng, L., Liu, D., Yu, Z. To be published
- Structure of the biotin carboxylase subunit of pyruvate carboxylase from Aquifex aeolicus at 2.2 A resolution. Kondo, S., Nakajima, Y., Sugio, S., Yong-Biao, J., Sueda, S. & Kondo, H. (2004). Acta Cryst. D60, 486-492
- Movement of the biotin carboxylase B-domain as a result of ATP binding. James B. Thoden, Carol Z. Blanchard, Hazel M. Holden and Grover L. Waldrop Movement of the biotin carboxylase B-domain as a result of ATP binding J Biol Chem. 2000 May 26;275(21):16183-90
Discovery
Contact us for a free confidential discussion
We help you reach your next milestone faster and more cost effectively
Contact us for a free confidential discussion