Discovery
Reposition existing drugs
Computational re-profiling is a highly efficient way of identifying and developing new opportunities for existing drug compounds. It is an excellent method of identifying alternative targets for candidate compounds and suggesting alternative compounds for known targets.
Computational re-profiling
Cresset Discovery offers a unique insight into the molecular characteristics behind biological activity using the XED force field. XED takes the 3D shape and electrostatics of molecules into account to produce a field view of compounds. This makes it possible to compare thousands of structures and fragments on the basis of biological similarity.
This approach has been successfully used to reposition drugs, and to extend the patents on compounds.
Computational chemistry experts from Cresset Discovery helped discover the repurposed compound RP0217 which has been patented by for a variety of disease indications
Identifying novel active structures
A drug repurposing company asked Cresset Discovery to identify an existing compound that was also active against PP2A that could be re-purposed as an anti-inflammatory.
The starting point for their search was the three ligands known to be active at PP2A, plus a crystal structure of PP2A bound to a large toxin that completely shut off its mechanism. Based on these Cresset’s approach was used to build a template for a compound that would be active at PP2A.
The molecular
The crystal structure was also imported into Forge and the bound ligand was manually trimmed back in order to identify the active pocket in the protein. This was used to create a fourth template for a screening run. The
Screening results in patented
The results of the four runs were evaluated and prioritized. A small number of compounds were purchased and screened. Within the screened compounds, one was found to inhibit PP2A with nanomolar activity. This existing compound is known internally as RP0217.
The initial hit was followed up with secondary screening, including ex vivo and in vivo models of inflammatory diseases. Through all of these validation experiments, RP0217 has been found to be an effective anti-inflammatory agent and has consequently been patented by Re-Pharm for a variety of disease indications.
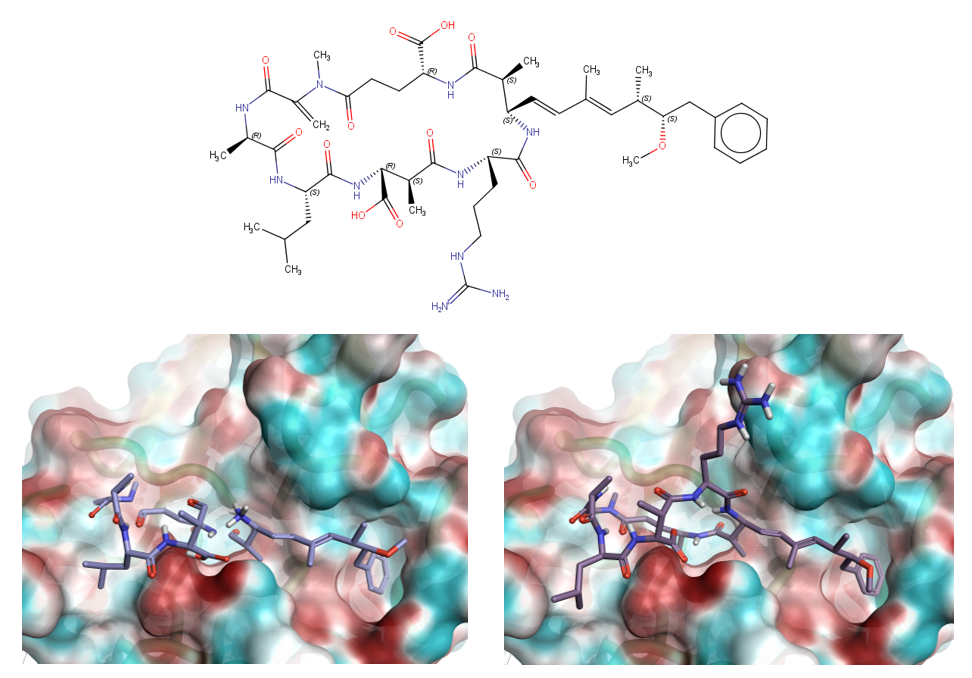
Top: Full structure of microcystin LR. Bottom left: Resolved regions of microcystin LR toxin from the 2ie3 crystal structure. Bottom right: Full ligand as modeled using Flare™ and Forge™ in the protein phosphatase 2A active site.
Discovery
Contact us for a free confidential discussion
We help you reach your next milestone faster and more cost effectively
Contact us for a free confidential discussion