In this case study we will use the new Spark docking feature of the Cresset bioisostere replacement and scaffold hopping tool to prioritize alternative R-groups picking ligand protein interactions directly from the active site of GSK-3β.
Introduction
GSK-3β is a Serine/Threonine kinase which participates to a variety of cellular processes; in particular, its widespread expression in adult brain suggests a fundamental role in neuronal signalling pathways. GSK-3β has also been linked to mood disorders such as bipolar disorders, depression, and schizophrenia, with a growing interest in the discovery of novel GSK-3β inhibitors as a potential treatment.
1H-indazole-3-carboxamides are a recently identified1,2 novel structural class of ATP-competitive GSK-3β inhibitors. Cmpd 1 (Figure 1 – left and Table 1) is a potent enzymatic and cellular GSK-3β inhibitor, also showing a favorable kinase selectivity profile, encouraging PK properties and efficacy in animal models of mania, but affected by hERG liability.
Lead optimization2 of Cmpd 1 to tackle hERG liability included the replacement of the (2-methoxyethyl)-4-methylpiperidine moiety with an oxanyl group to reduce the compound basicity, and the replacement of the di-F-phenyl moiety with more polar groups to help reduce lipophilicity. This effort led to the identification of Cmpd 14 (Figure 1 – right and Table 1), showing slightly higher GSK-3β activity and significantly reduced hERG liability.
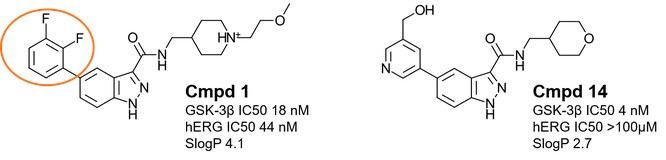
Figure 1. Left: Cmpd1 is a potent and kinase selective GSK-3β inhibitor affected by hERG liability. Right: The optimized Cmpd 14 shows enhanced GSK-3β activity and reduced hERG liability.
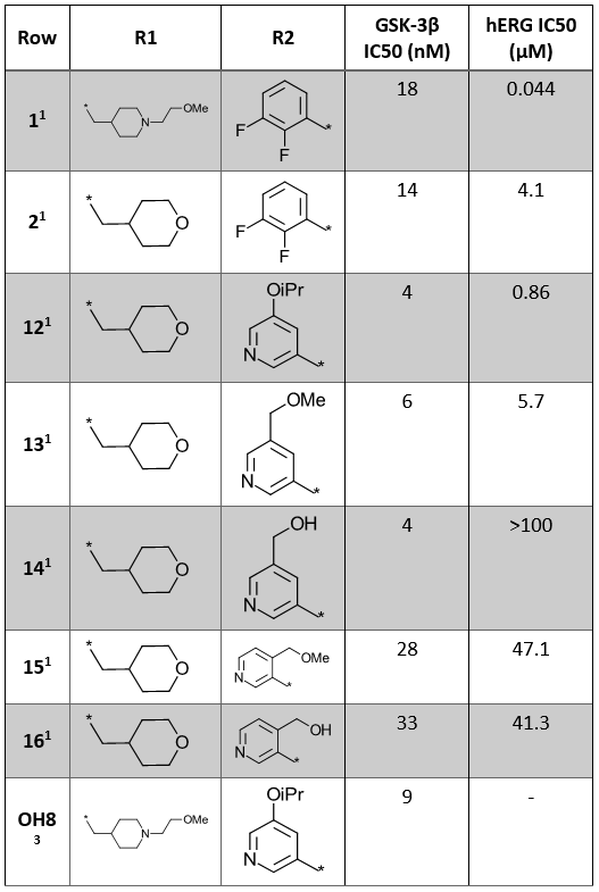
Further SAR data for analogues of Cmpd 1 is shown in Table 1.
Table 1. SAR exploration of R1 and R2 positions of the 1H-indazole-3-carboxamide core.1,3
Here we will use the ‘docking’ feature in Spark to rapidly identify alternative R-groups to the di-F-phenyl moiety (orange circle in Figure 1 – left). This method can be used to grow ligands and fragments into unoccupied pockets of the target protein and to find novel results making interactions with the active site of the protein not mapped by an existing starter or reference molecule. More specifically, we will search for alternative R-groups making direct interactions with the GSK-3β active site to enhance GSK-3β activity, and less lipophilic to reduce the likelihood of hERG liability.
Protein-ligand complex
When using the docking method in Spark, it is highly recommended to start the experiment from an accurately prepared protein, in order to add missing hydrogen atoms, optimize the internal hydrogen bond network, remove atomic clashes and assign optimal protonation states. The preparation of the protein-ligand complex of Cmpd 1 bound to GSK-3β (PDB: 6TCU) was performed using Flare™, the Cresset structure-based design platform.
Visual inspection of protein-ligand contacts (Figure 2) shows that the indazole carboxamide scaffold interacts with the hinge domain of GSK-3β by means of three H-bond interactions with the backbone carbonyl group of Asp133, the backbone NH and the backbone carbonyl of Val135. The piperidine moiety is totally exposed to the solvent, while the di-F-phenyl ring is in proximity of catalytic Lys85.
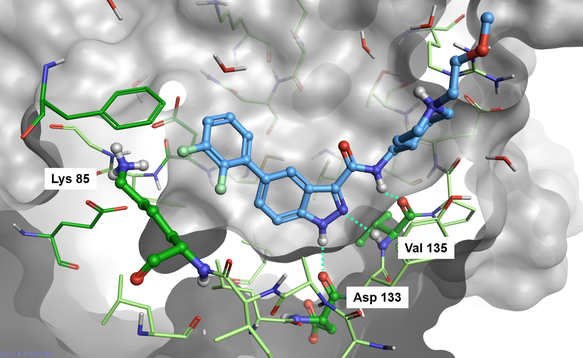
Figure 2. Cmpd1 bound to the GSK-3β active site (PDB: 6TCU).
Docking in Spark
The docking in Spark workflow starts by removing the portion of the starter molecules which you wish to replace (Figure 3, A-B). A new fragment is then attached to the truncated starter molecule and placed in the active site of the protein in a sensible orientation guided by the protein electrostatics calculated with the Cresset XED force field (Figure 1, C). The pose of the new result molecule is then optimized by using the Lead Finder™ ‘score only’ docking mode (Figure 1, D). Finally, the best results are prioritized by means of the Lead Finder ‘Rank’ scoring function, optimized to correctly reproduce the crystallographic pose of known ligands, and sorted in order of ascending Docking Score in the Spark results table (the more negative the score, the better).
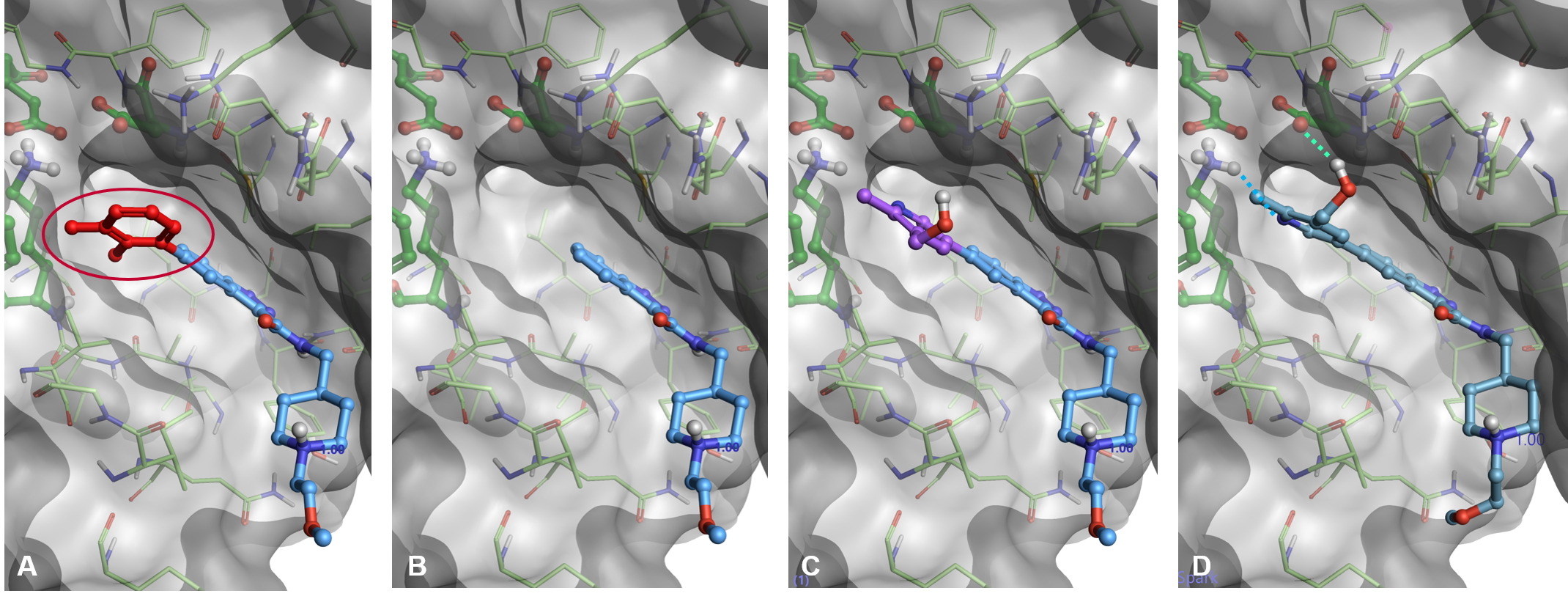
Figure 3. Docking workflow in Spark. A) A portion of the starter molecule is selected for replacement. B): The selected fragment is deleted. C): A new fragment is attached in a sensible orientation to the truncated starter molecule. D): The pose of the new result molecule is optimized using Lead Finder.
We used the ‘Scaffold Hopping or R-group Replacement’ wizard (Figure 4 - left), which provides an easy-to-follow interface to the set-up of the experiment. The 6TCU protein-ligand complex prepared with Flare was imported in Spark using the input protonation state, choosing Cmpd 1 as the starter molecule, and deleting all crystallographic water molecules. The di-F-phenyl moiety was selected as the region of the starter molecule to replace (Figure 4 -right).
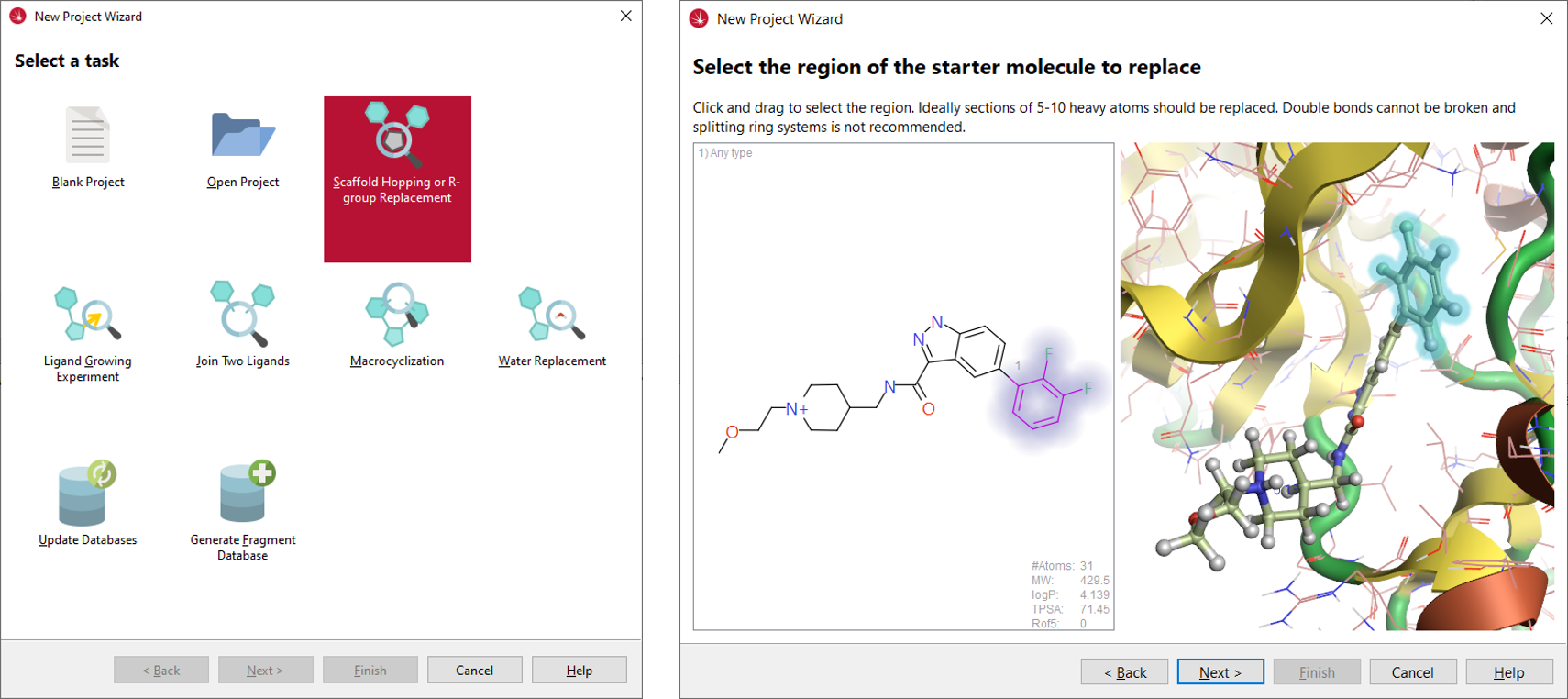
Figure 4. Left: the new project wizard in Spark. Right: in the Spark wizard, the region of the starter molecule selected for replacement is highlighted by halos.
On exiting the wizard, the Spark calculation method was set to ‘Docking’. As this method is more computationally demanding than traditional Spark searches based on ligand similarity, it is recommended to set appropriate physico-chemical filters to limit the search space and reduce the calculation time, focussing the experiment on the desired results. Using the properties of the starter molecule (MW 429.5, SlogP 4.1) as a guide, the molecular weight of the potential results was set to be lower than 466, and SlogP to be lower than 3.5. Substituted phenyls and 2-hydroxy-pyridines were also filtered out from the list of potential results using an appropriate SMARTS filter (Figure 5).
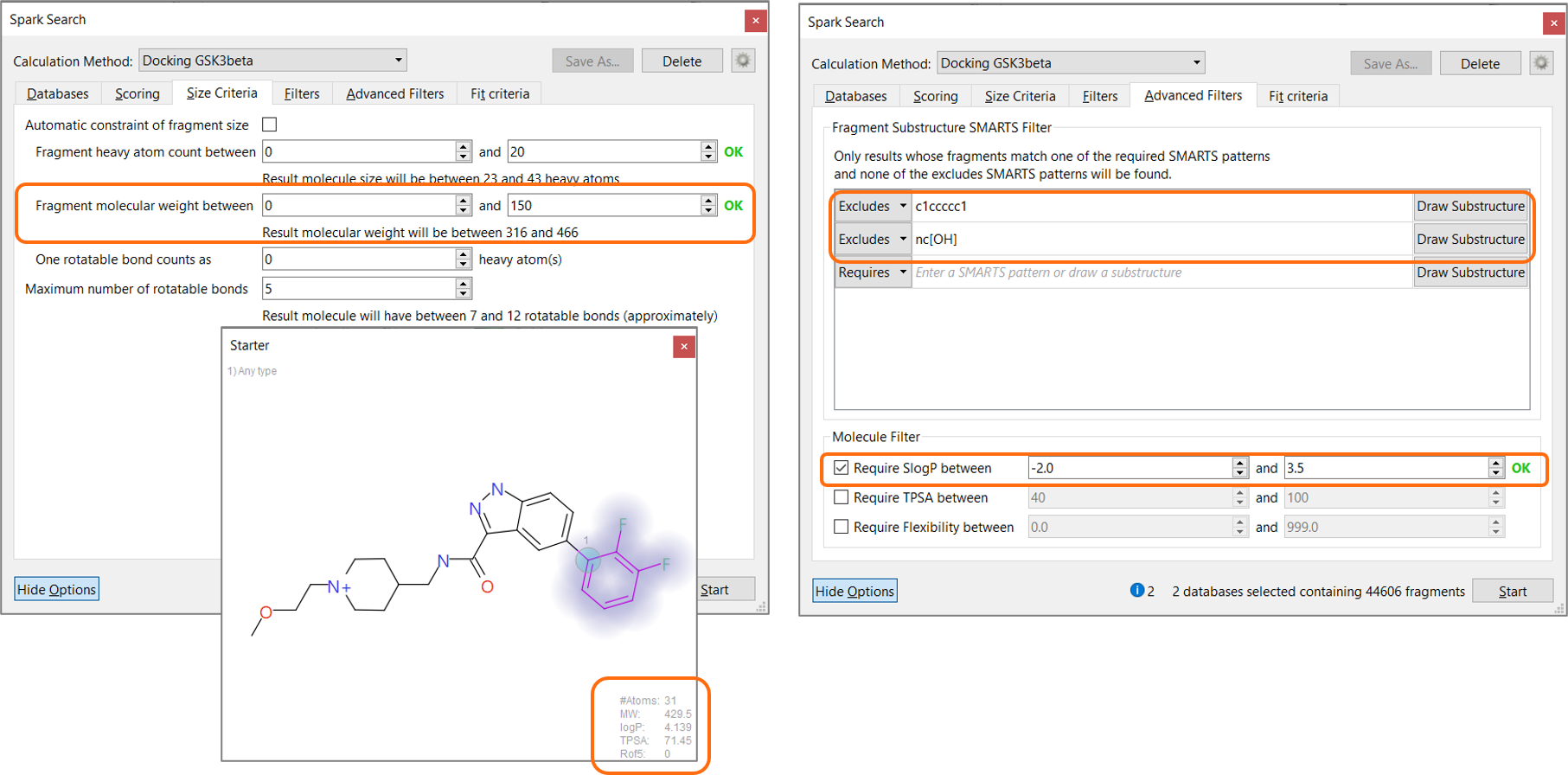
Figure 5. Setting appropriate physico-chemical filters during the Spark search focuses the experiment on the desired results and reduces calculation time.
The experiment was run on a database of 4.5K aromatic boronic acids and 40K aromatic halides derived from eMolecules building blocks4 to closely replicate the chemistry used in the original publication.2
Results
The docking in Spark experiment correctly identifies pyridin-3-yl substituents among the top-scoring clusters of results (Figure 6, left), including R-groups identical or similar (Figure 6 – middle) to those reported in Table 1. Among the top scoring Spark results we can also find a variety of bicyclic heterocycles with limited molecular weight and lipophilicity, which might provide interesting alternatives to pyridin-3-yl substituents for a further exploration of this lead series. Some examples are shown in Figure 6, right.
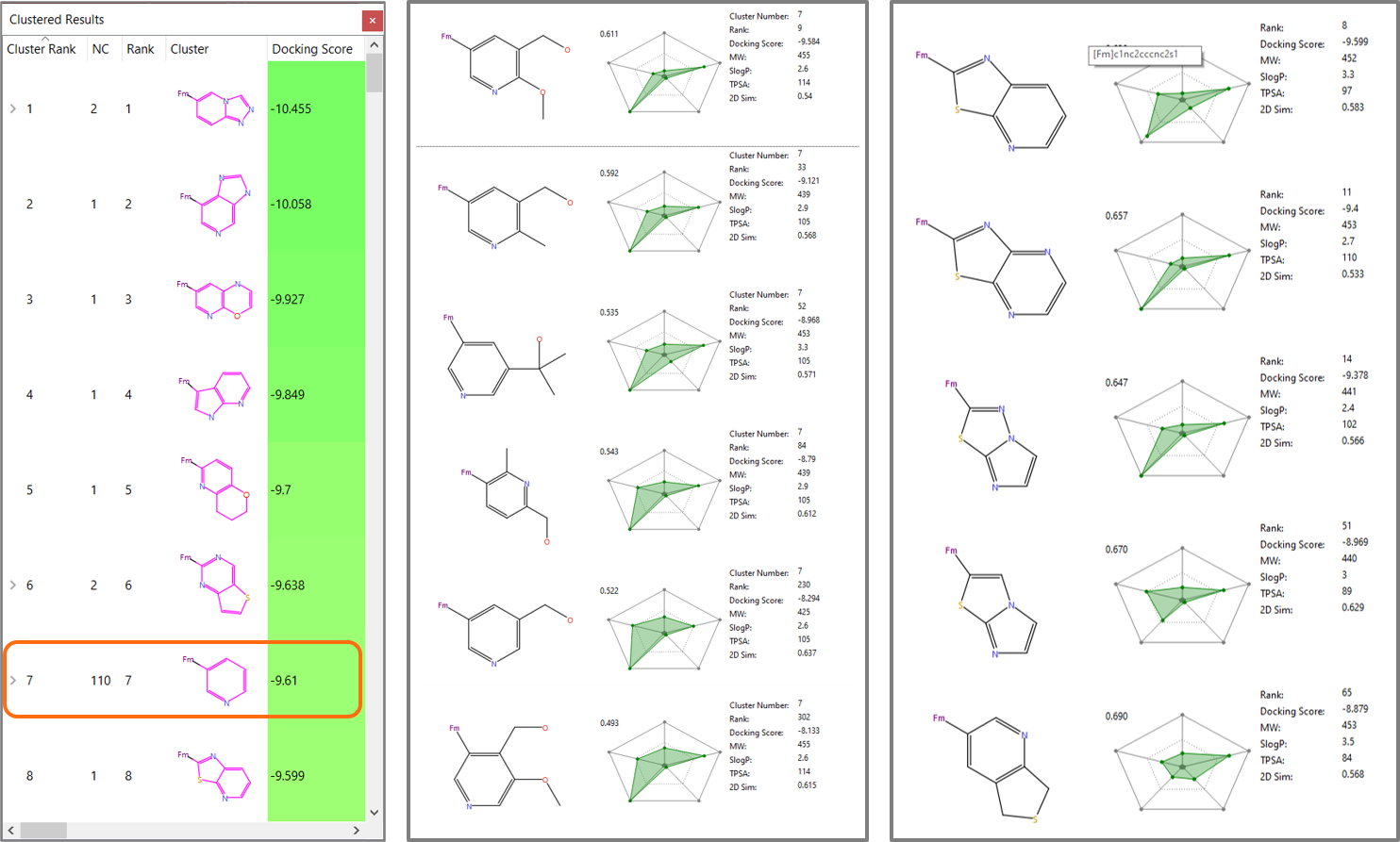
Figure 6. Left: Pyridin-3-yl substituents2 have cluster rank 7 in the Spark results. Middle: Members of the pyridin-3-yl substituents cluster similar to those in Table 1. Right: bicyclic heterocycles with limited molecular weight and lipophilicity found by Spark.
Interestingly the binding mode proposed by Spark for many members of the pyridin-3-yl cluster involves an H-bond interaction with Lys85 and in some cases an aromatic-aromatic interaction with Phe67, consistent with the crystallographic binding mode of compound OH84 (PDB: 6Y9R, Figure 7). Not surprisingly though, the docking method tends to favour 3-pyridines carrying substituents (such as hydroxyl) making additional H-bond interactions with residues lining this region of the active site, such as Asp 200.
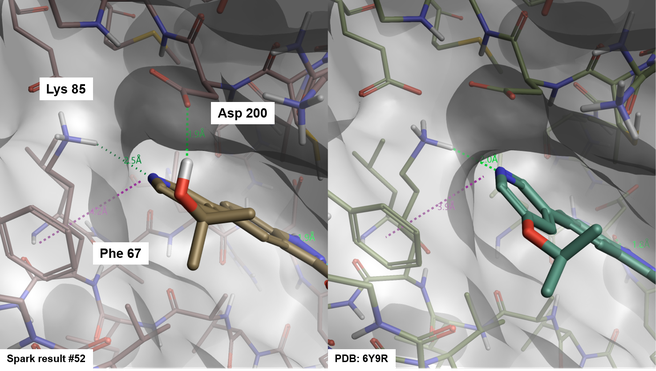
Figure 7. Binding mode of Spark result #52 (left) and compound OH8 from PDB: 6Y9R (right).
Conclusion
The ‘docking’ feature enables Spark to find fragments picking ligand-protein interactions directly from the protein active site. This method can be used to grow ligands and fragments into unoccupied pockets of the target protein, and to find novel results making interactions with the active site of your protein not mapped by an existing starter or reference molecule.
References
1. Ombrato R. et al., J. Chem. Inf. Model. 2015, 55, 2540−2551
2. Prati F. et al., ACS Med. Chem. Lett. 2020, 11, 825−831
3. Buonfiglio R. et al., Molecules 2020, 25(9), 2163
4. https://www.emolecules.com/info/products-building-blocks.html