Jenny Brookes†, Tim Cheeseright†, Mark Mackey†
†Cresset, New Cambridge House, Bassingbourn Road, Litlington, Cambridgeshire, SG8 0SS, UK
Abstract
This case study demonstrates a workflow which uses positional analogue scanning combined with free energy calculations to rapidly explore the SAR landscape around an individual hit compound, accelerating the hit-to-lead process. We use the new Hit Expander module within Flare™, Cresset’s software for ligand and structure-based design, to generate a range of suggestions around a starting hit, and then show that triaging these suggestions using Flare Free Energy Perturbation (FEP) both retrieves known highly active molecules and makes sensible suggestions for further modifications for improved activity.
Introduction
Lead discovery & optimization is a lengthy and expensive process affected by a very high attrition rate. Hundreds of compounds are frequently synthesized and tested, with an enormous monetary, material, and time expenditure in lab-based activities. Using smart computational methods can significantly help streamline and speed up this process, by efficiently prioritizing the best molecules to progress.
In this case study we demonstrate how easy to use, fast methods such as Hit Expander, combined with accurate predictions of activity using Flare FEP, can be used to reliably and rapidly discover promising lead compounds that can be taken forward with confidence to experimental testing, saving time and money. We show success even in the case here where relevant crystallographic data on the lead series of interest is not available.
This study uses published data taken from US Patent US9796708,1 describing the invention of a class of azaindole Cyclin-Dependent Kinase 9 (CDK9) inhibitors (Figure 1), and from a follow-up paper from Tong et al.2 describing the lead optimization campaign of the compounds in Figure 2. Inhibitors of CDK9 are attractive as they promise potential anti-cancer, cardiology and virology therapeutic applications.3-5 Studies also suggest they have uses in neuropathic, chronic and inflammatory-related pain treatment.6-7
From the collection of over 1,000 inhibitors reported in the patent, we chose just 2 known actives: one was used as a ‘reference’ in setting up the FEP calculation and the other was treated as an initial ‘hit’ compound to create 84 variations with Hit Expander.
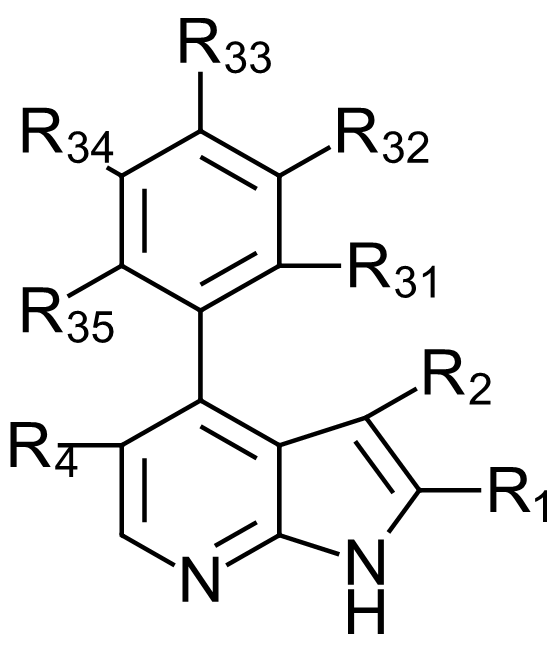 Figure 1. General formula of the azaindole CDK9 inhibitors reported in US9796708.
Figure 1. General formula of the azaindole CDK9 inhibitors reported in US9796708.
Modelling the starting ligand-protein complexes
Compound 1 (Figure 2 – left) is a potent inhibitor of CDK9 (US9796708, IC50 = 14 nM /ΔGbinding = -10.7 kcal/mol) and exhibits more than fifty-fold selectivity over CDK1, 2 and 7. However, at the time of writing, there was no available co-crystal X-ray structure for CDK9 bound to this compound (or for a close analogue). Accordingly, the binding pose of compound 1 (4PEP) within the CDK9 active site was modelled starting from the crystallographic pose of a similar ligand bound to the active site of CDK2 (PDB: 7M2F), and used as the starting point for the in silico lead optimization campaign.
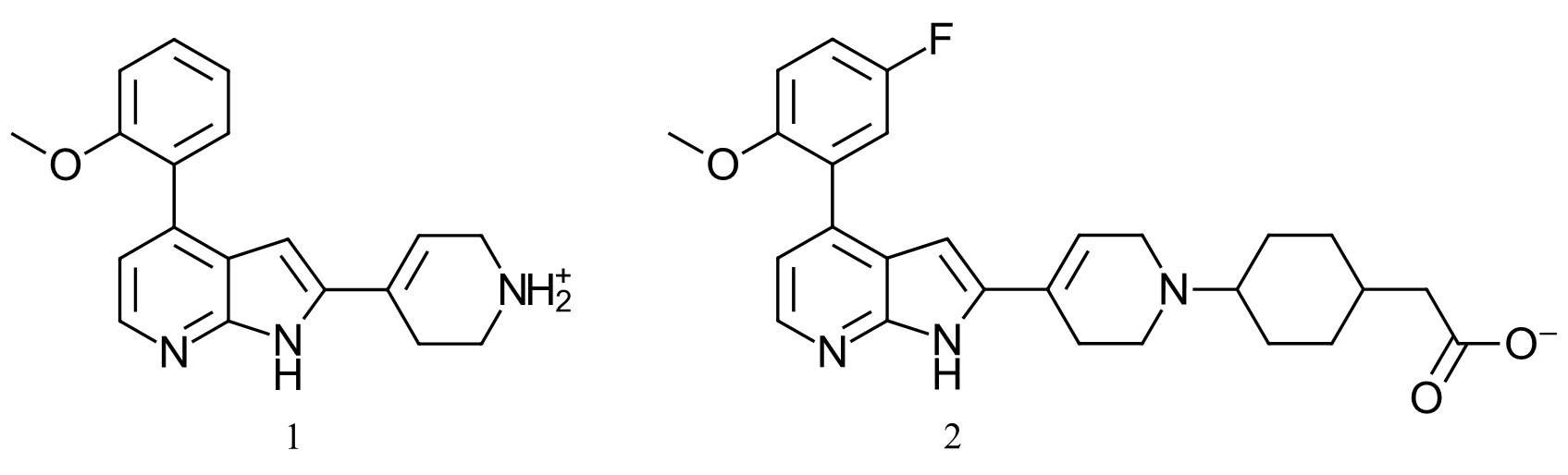 Figure 2, Compound ‘1’ referred as 4PEP in this case study is a compound from Tong et al1 patent (US9796708)[GT1] , with an IC50 of 14nM for CDK9. Compound 2 is the crystallographic ligand bound to the CDK2 PDB:7M2F structure, which was used as a reference to align 4PEP.
Figure 2, Compound ‘1’ referred as 4PEP in this case study is a compound from Tong et al1 patent (US9796708)[GT1] , with an IC50 of 14nM for CDK9. Compound 2 is the crystallographic ligand bound to the CDK2 PDB:7M2F structure, which was used as a reference to align 4PEP.
A CDK9 protein structure with a potent inhibitor bound (IC50 = 11nM/-10.9 kcal/mol) was identified that bound in a similar manner, (PDB:6GZH). This protein was superimposed onto the CDK2 structure. The superimposition was inspected and confirmed to be well aligned, and that the ligands shared a similar binding mode in the active sites. Then the 4PEP ligand was aligned in Flare using the Conf Hunt & Align ligand alignment substructure method, to generate a relevant binding pose for both the aligned proteins. The CDK9 protein is the one of interest for this work and to evaluate the stability of the modelled binding pose of 4PEP in the active site a 20ns molecular dynamics simulation was undertaken of 4PEP in the prepared CDK9 protein (PDB:6GZH).
Preliminary Molecular Dynamics studies
Running Molecular Dynamics (MD) as a precursor to FEP is always a useful validation of the protein-ligand complex set-up, which allows the user to check that:
- The binding mode of the ligands of interest is preserved during the simulation.
- The active site residues near the ligands are set-up correctly in terms of charge, tautomer and rotamer state.
- Protein-ligand and protein-water interactions are occurring as expected (i.e., in this case we expect binding to the hinge region of CDK9).
MD studies can also help determine whether you may need to include in your model dimeric/ multimeric proteins and any other molecules that potentially aid stability. Such studies have established that in cyclin kinases, the cyclin should generally be included. Accordingly for this case study, as CDK9 is a heterodimeric complex, the dimer (consisting of the kinase and the cyclin) was modelled in MD and consequently when using Flare FEP.
Water analysis with 3D-RISM
Given the lack of crystal data for compound 2 in CDK9, there was added impetus to do some careful water analysis at the binding site. A stable snapshot from the end of the MD run was taken and then used to run 3D-RISM analysis in Flare on these coordinates. This experiment used the Cresset XED force field8 with advanced inter-molecular descriptions to place waters in the active site whilst the ligand is present (Figure 3). The prepared protein snapshot with all the water positions predicted by 3D-RISM was then used in the subsequent Flare FEP calculations.
The use of 3D-RISM to analyse water positions can be crucially important because a missing water from the ‘right’ place (in a buried position or making ligand/protein/other interactions) or a misplaced water in the ‘wrong’ place can cause large energetic discrepancies and introduce anomalies in FEP. Water analysis with 3D-RISM using XED force field can provide accurate and rapid determination of likely bound water locations: given its simplicity and very modest computational cost (minutes on 1-4 local CPUs), it is worth running this experiment before going forward with more costly methods (such as FEP).
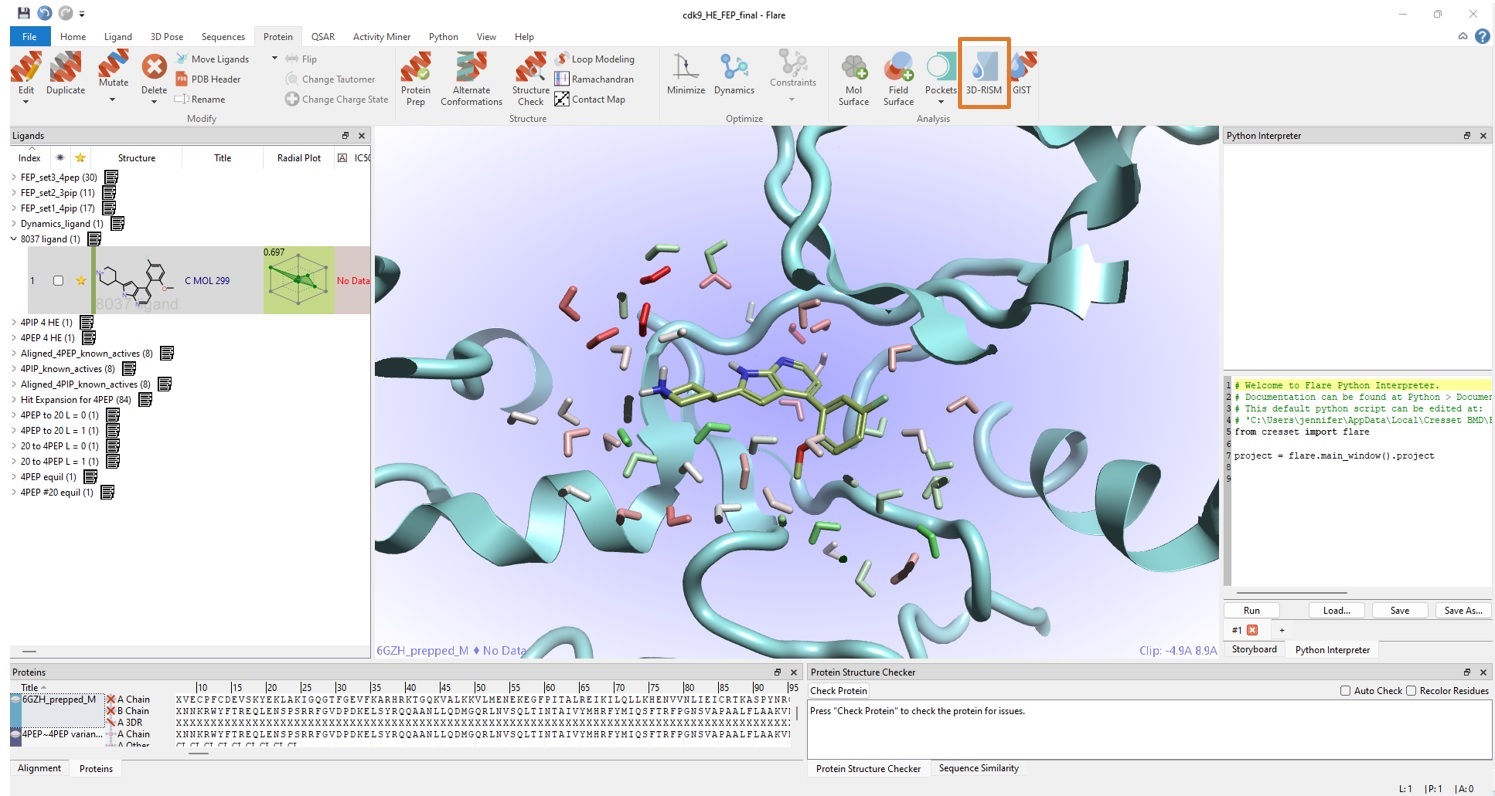 Figure 3. 3D-RISM in Flare can be used to ‘discover’ likely bound water sites, providing valuable input to Flare FEP simulations. The colouring (red to green) of predicted water sites corresponds to unfavourable to favourable ΔG, respectively.
Figure 3. 3D-RISM in Flare can be used to ‘discover’ likely bound water sites, providing valuable input to Flare FEP simulations. The colouring (red to green) of predicted water sites corresponds to unfavourable to favourable ΔG, respectively.
Hit Expander
Flare’s Hit Expander employs the concept of making small changes to a hit molecule via positional analogue scanning (PAS),9 resulting in many design ideas in a congeneric series. For this case study, we chose a known active from US97967081 as the hit to expand. This compound, which we will call ‘4PEP’ (Figure 2 in 2D or Figure 4 – left in 3D), shows minimal inhibitory concentration with CDK9 IC50 = 24 nM/ΔGbinding = -10.4 kcal/mol and cellular antiproliferative activity (H929 IC50 = 3000 nM/ ΔG = -7.5 kcal/mol).
In preparation to the Hit Expander experiment, 4PEP was aligned onto the prepared system of the CDK9/compound 2 complex, using the same snapshot from MD which was used for the 3D-RISM experiment.
In Hit Expander you can choose any part of the core in your molecule that you would like to expand. We selected the whole ligand structure and applied the default settings (Figure 4 – middle) which will add substituents to all substitutable positions throughout the structure, by performing:
- Single point substitutions Me, F, Cl, OH, OMe
- Aromatic substitutions C > N, N > C, N > O and O > N
Other options are available from the Hit Expander panel, but this was more than sufficient to generate a largish set of 84 hit variants (Figure 4 – right) and rapidly and more than adequately explore the immediate chemical landscape around the hit compound (as will be shown in the ‘Results’ section). After generating all the possible variations of the initial hit, these are deduplicated and gently minimized to generate optimized 3D structures in preparation to further studies.
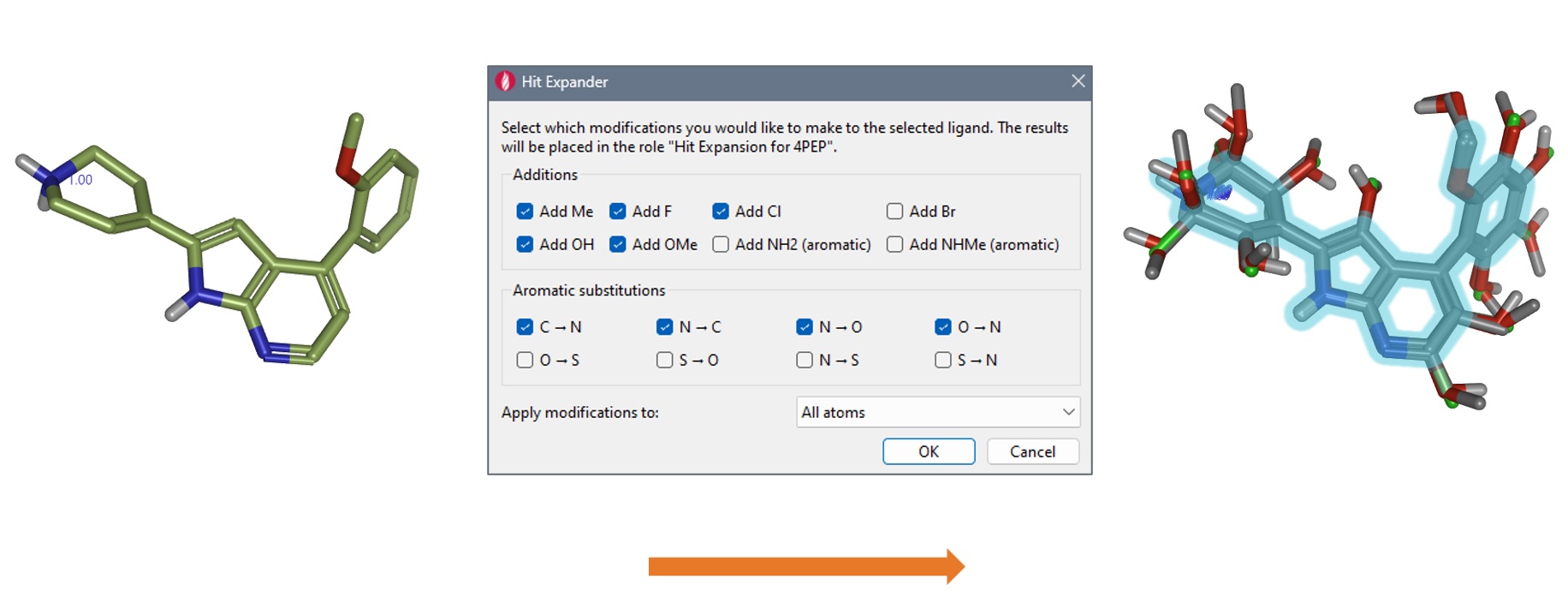 Figure 4. Using Hit Expander (middle) on the '4PEP' hit compound (left), to quickly generate promising candidates. In this case, 84 new molecules were created (right).
Figure 4. Using Hit Expander (middle) on the '4PEP' hit compound (left), to quickly generate promising candidates. In this case, 84 new molecules were created (right).
Flare FEP
Relative FEP is particularly suitable for triaging the hits generated by Hit Expander because the suggested molecules have only small structural changes from the reference molecule, and FEP calculations tend to be more reliable for small perturbations. All Flare FEP runs reported here use the newest available (as of writing) OpenFF2.0 small molecule forcefield10-11 combined with custom torsions generated using a semiempirical tight-binding method GFN2-xTB12-13 that approximates to QM, together with the protein forcefield AMBER ff14SB14.
First Flare FEP experiment: minimum spanning tree, production mode
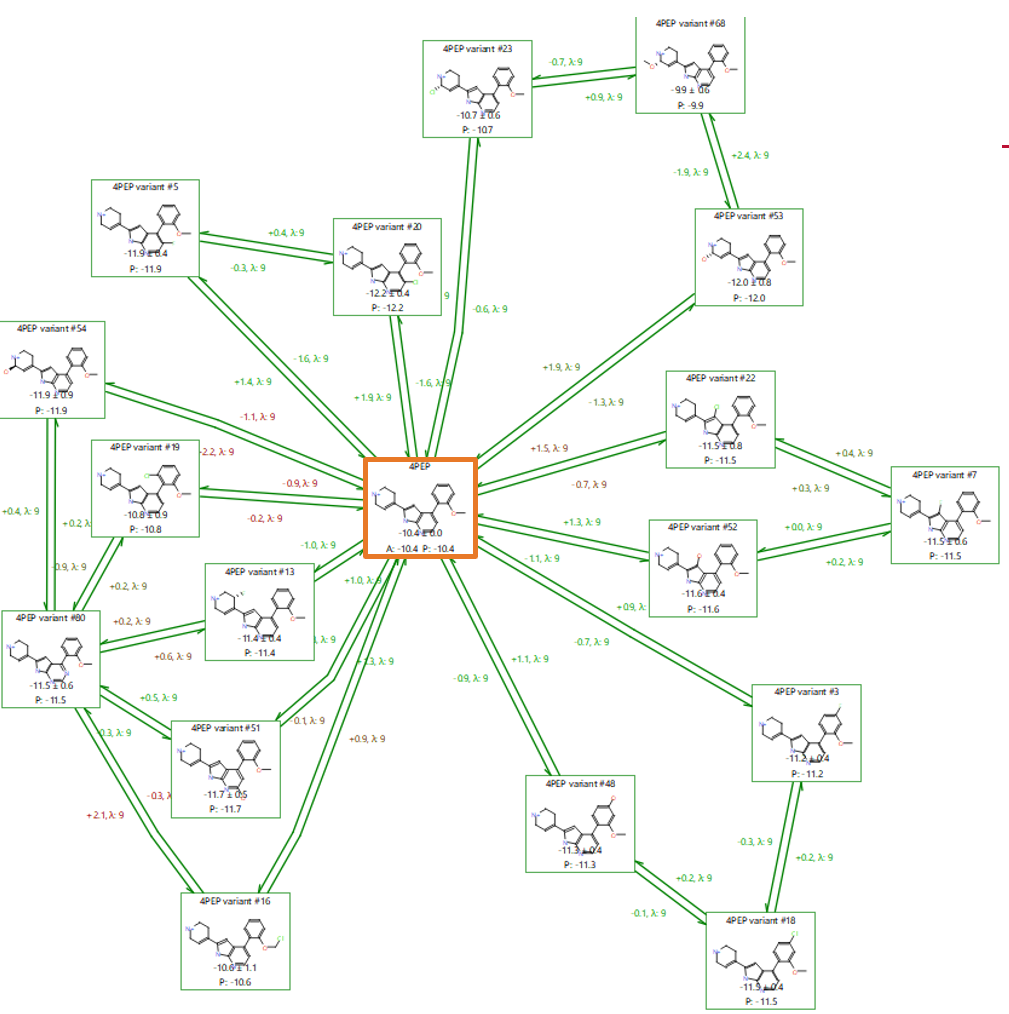
The set of 84 variations generated by Hit Expanders are already very closely aligned given only small changes were made incrementally to 4PEP. Therefore a ‘minimum spanning tree’ perturbation graph based on the known 4PEP compound and the 84 suggestions (Figure 5) is particularly suitable for a first triage.
A minimum spanning tree is an efficient graph with no cycles, that maximises links to the centre where possible and branches off from the first link from the centre where it is not. In particular, setting up the network using Flare’s ‘production’ mode biases the graph to minimise connection distances to the known active (Figure 5). All these perturbations have good ‘link scores’15 (> 0.6) and so are likely to provide reliable predictions, reducing the need to calculate multiple (redundant) connections. For this reason, it is possible to run the calculation with the ‘Very Quick’ mode in Flare FEP, using unidirectional, ‘single mode’ perturbations (from the centre outwards), and 1ns sampling time.
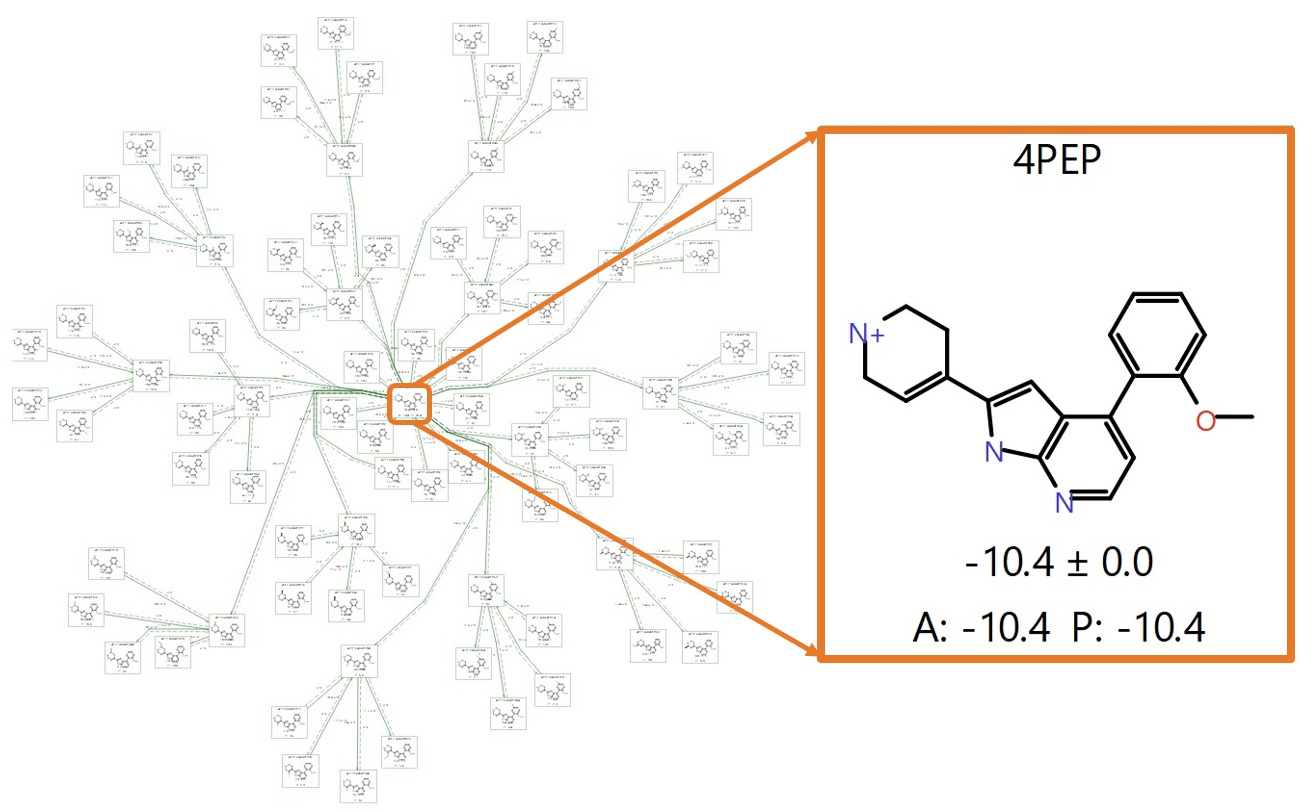 Figure 5. A completed production run using a ‘minimum spanning tree’ map with 4PEP (known active) at the centre, highlighted in the orange box. ‘A’ denotes the Actual ΔG and ‘P’ the Predicted ΔG (in kcal/mol).
Figure 5. A completed production run using a ‘minimum spanning tree’ map with 4PEP (known active) at the centre, highlighted in the orange box. ‘A’ denotes the Actual ΔG and ‘P’ the Predicted ΔG (in kcal/mol).
This first Flare FEP run counts 85 ligands in total, including the known active 4PEP at the centre and making predictions for 84 ligands, so the graph consisted of 84 links and 84 perturbations for a total of 756 Lambda windows (assuming each perturbation requires the default 9 Lambda windows. In practice, Flare FEP implements an adaptive Lambda sampling so the Lambda windows required for convergence are determined in a fully automated fashion).
Second Flare FEP experiment: normal (multi-connected) graph, production mode
The second FEP experiment was run to refine the results of the first experiment by taking forward the 17 top predicted binders (20% of the total). The known active 4PEP was again used as the centre of a ‘normal’ graph (multi-connected / with cycles, Figure 6), creating a graph with 25 links, 50 perturbations and 450 Lambda windows. This second calculation was also run in production mode, but calculating both directions of perturbation, as this gives a larger degree of error analysis and enhances overall accuracy.
Both runs together can be run comfortably overnight on a small cluster of 10 or so GPUs. Using only 1ns sampling time per Lambda window can be sufficient in cases like this with small changes in your ligand data set: furthermore, the sampling time can be easily extended (Flare FEP default is 4 ns) where needed and this can be done post-calculation on a link-by-link basis (depending on where it might be needed).
Figure 6. A completed Flare FEP production run using a ‘normal’ connected map with 4PEP (known active) at the centre, highlighted in the orange box.
Results
Normal connected graphs, such as the one in Figure 7, allow the user to do a fast triage of the results by cycle closure error analysis. Examining the cycles in Figure 7 showed larger cycle closure errors for some compounds (circled in orange), and so the predictions for these compounds should be treated as being less reliable.
Two of the ligands in Figure 7 (boxes, purple) which were predicted to be good binders by Hit Expander and Flare FEP are indeed known to be good binders. ‘4PEP variant #3’ was predicted to have a binding free energy of -11.2 kcal/mol and is reported in US9796708 to have an experimental binding of 16 nM (ΔGbinding -10.6 kcal/mol). ‘4PEP variant #18’ was predicted to have a binding energy of -11.5 kcal/mol, which again compares favourably to the experimental binding constant of 28nM (ΔGbinding -10.3 kcal/mol). This provides confidence that the predictions for the other compounds are also likely to be accurate.
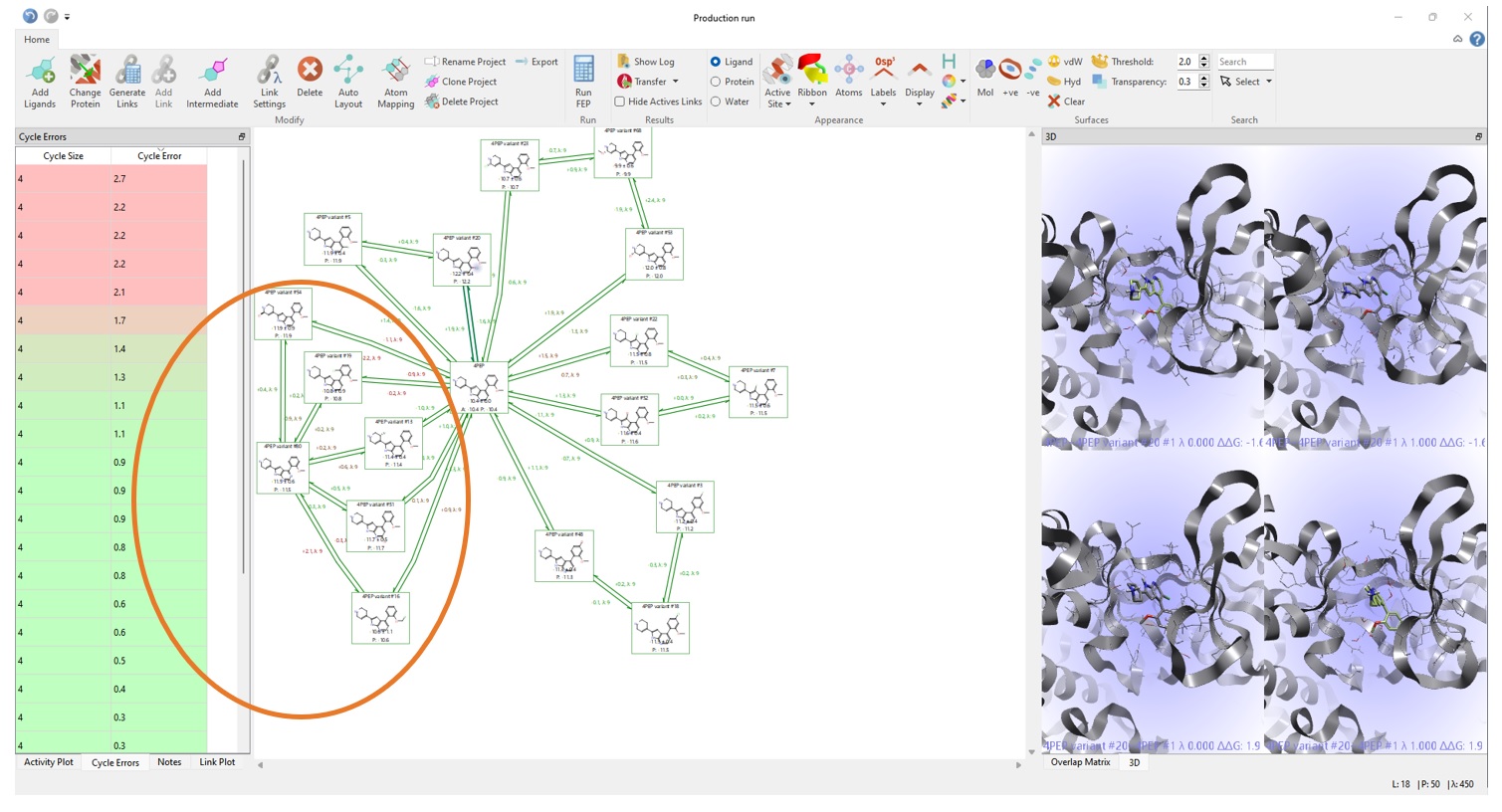 Figure 7. Examining the final FEP results in the Flare FEP project space.
Figure 7. Examining the final FEP results in the Flare FEP project space.
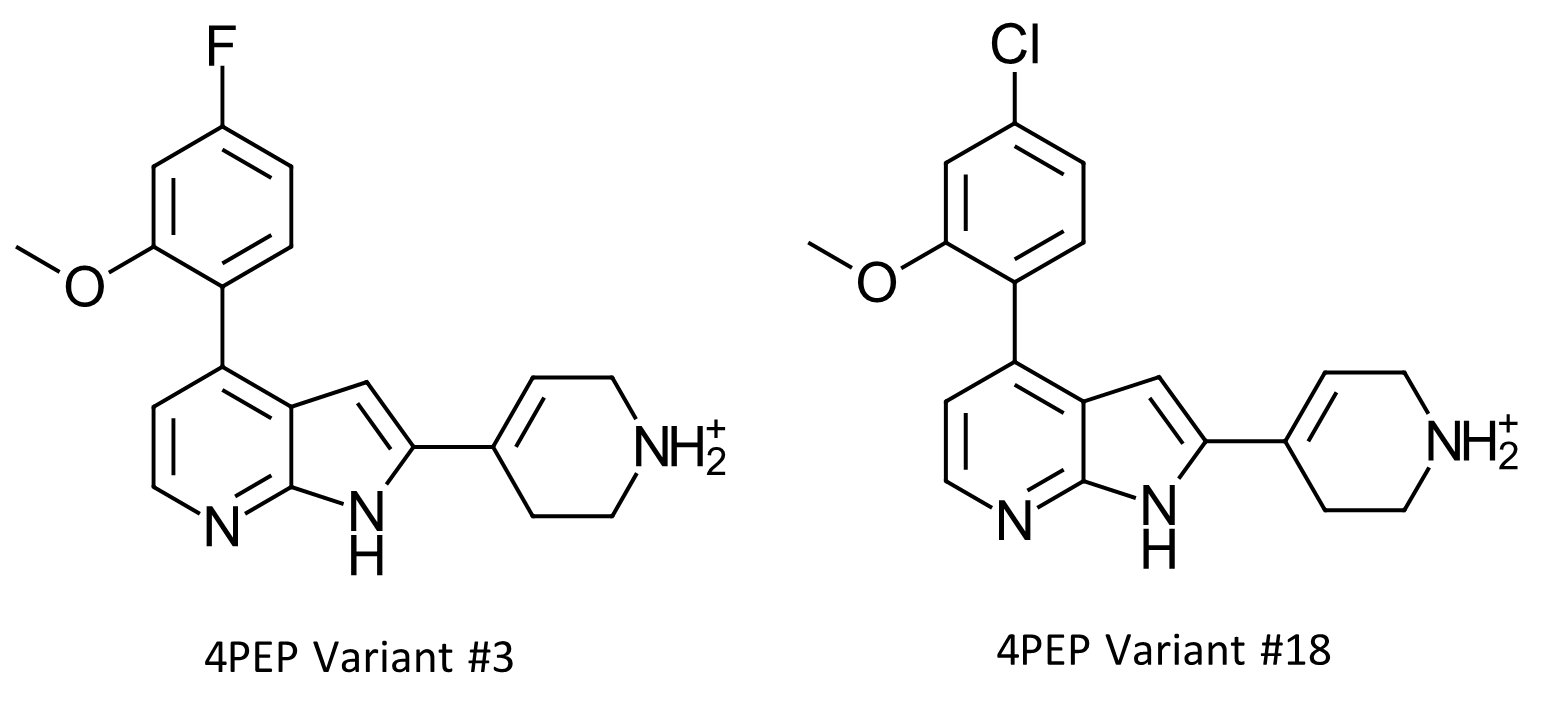 Figure 8. Two designs from Hit Expander and Flare FEP are compounds from patent US9796708 and have measured activity data. Variant #3 has a predicted ΔG value of -11.2kcal/mol and a measured IC50 of 16nM (ΔGbinding -10.6 kcal/mol). Variant #18 has a predicted ΔG value of -11.5 kcal.mol, and an experimental IC50 value of 28nM (ΔGbinding -10.3 kcal/mol).
Figure 8. Two designs from Hit Expander and Flare FEP are compounds from patent US9796708 and have measured activity data. Variant #3 has a predicted ΔG value of -11.2kcal/mol and a measured IC50 of 16nM (ΔGbinding -10.6 kcal/mol). Variant #18 has a predicted ΔG value of -11.5 kcal.mol, and an experimental IC50 value of 28nM (ΔGbinding -10.3 kcal/mol).
Excluding the above points, the three remaining cycles in the graph of Figure 7 show good overlap, little hysteresis and good cycle errors, so we could take forward novel suggestions from any of these 8 possibilities. However, we will focus on just on the cycle of this set shown in Figure 9, which includes the ligand with the best (more negative) predicted ΔGbinding combined with the lowest error and is the top ranking binder from the 17 predictions: ‘4PEP variant #20’ (in the blue box).
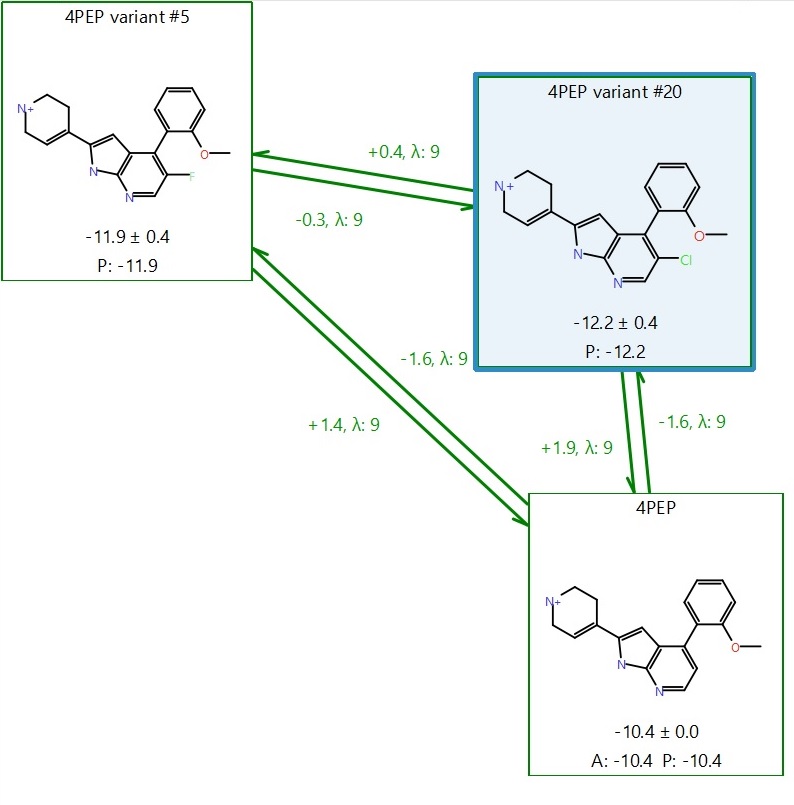 Figure 9. Result to take forward for further investigation: our lead ‘4PEP variant #20’ in the blue box.
Figure 9. Result to take forward for further investigation: our lead ‘4PEP variant #20’ in the blue box.
Figure 9 shows a cycle with high activity predictions, low error, no hysteresis between forward and backward perturbations, overall cycle closure error ΔΔG ~ 0 in both clockwise and anti-clockwise directions, and good overlap in the phase space matrices. We can reliably pick a result from here and go for ‘4PEP variant #20’ as a potential lead.
With a predicted ΔGbinding = -12.2 +/- 0.4 kcal/mol, #20 is expected to be 1.8 kcal/mol better at binding than 4PEP (ΔGbinding = -10.4 kcal/mol), which satisfies the assumption that if the ΔG in the FEP calculation is predicted to be at least 1.5-2 kcal/mol lower than the reference, then the predicted ligand should be at least as active as the reference, or more active.
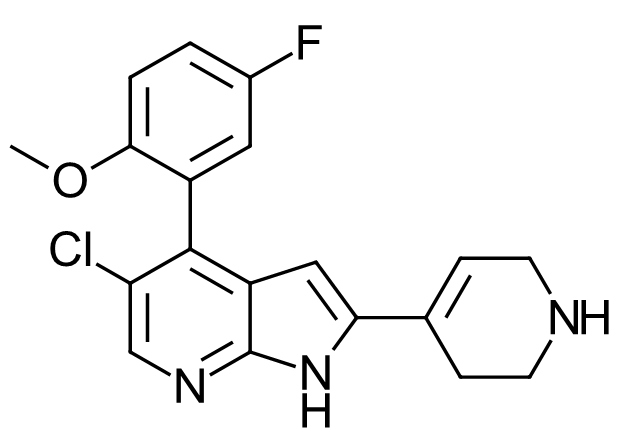 Figure 10. Compound 662 from US9796708 patent. Measured CDK9 IC50 = 28nM. This compound is 5-Fluoro derivative of 4PEP variant #20.
Figure 10. Compound 662 from US9796708 patent. Measured CDK9 IC50 = 28nM. This compound is 5-Fluoro derivative of 4PEP variant #20.
Whilst #20 does not appear in the patent data, a similar compound, only differing from #20 by an extra fluorine substitution opposite the methoxy group, does (Figure 10, US9796708, Example 662). This compound demonstrates a very slightly lower CDK9 activity than 4PEP (CDK9 IC50 = 28nM /ΔGbinding -10.3 kcal/mol versus 24nM /-10.4 kcal/mol for 4PEP) however has a better cellular antiproliferative activity (H929 IC50 = 83nM /-9.6 kcal/mol versus 3000nM / -7.5 kcal/mol for 4PEP).
There is a similar trend for other variants with chlorine substitutions a the 5-position (as in #20) described in the patent, that have improved cell viability, likely due to increase lipophilicity. So our prediction of #20, found via this workflow, holds promise of good CDK9 binding affinity combined with possibly improving the whole cell data.
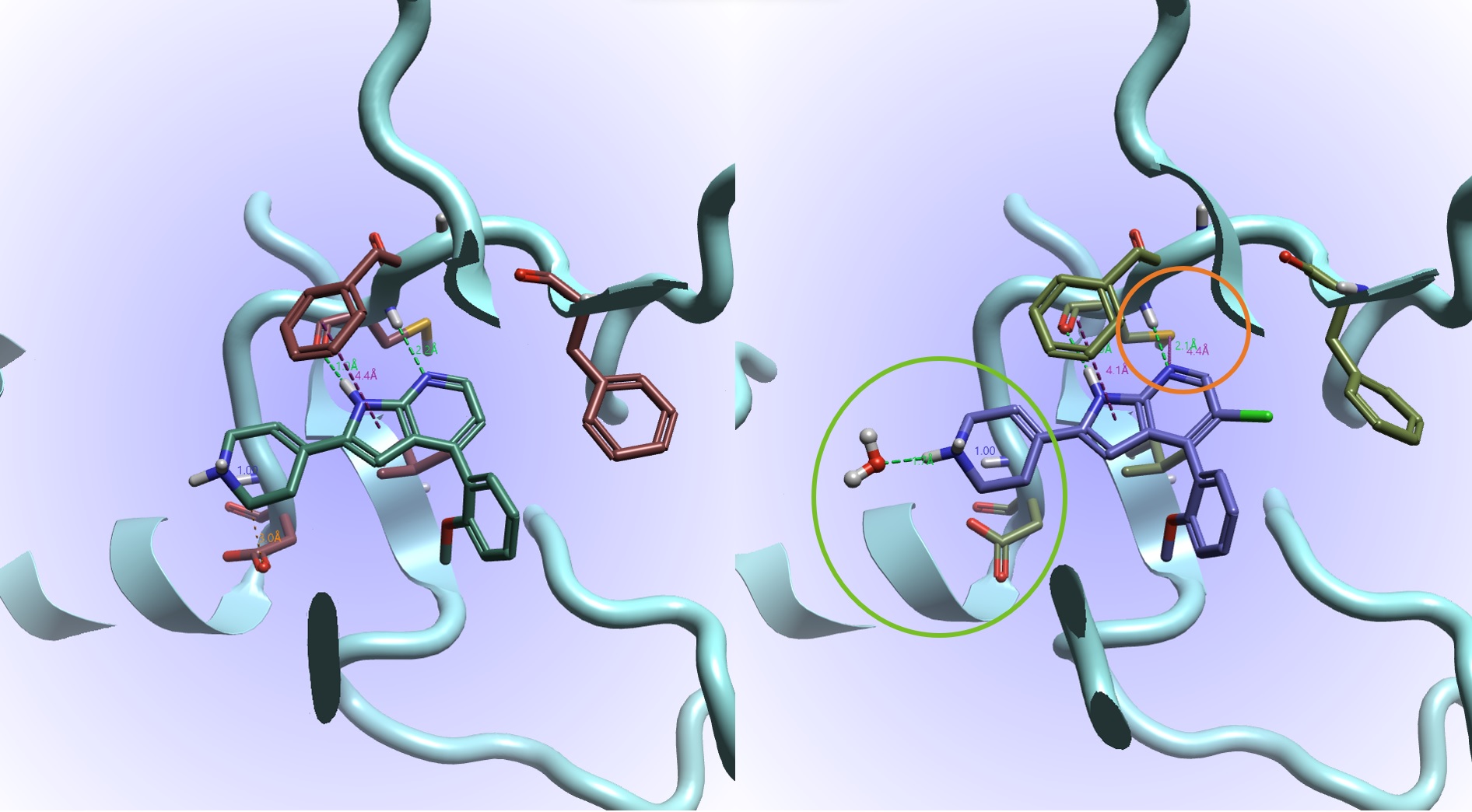 Figure 11. Comparing equilibrated 4PEP (left) and #20 (right) from the Flare FEP result. Shown here are the interactions: hydrogen bonds (green), steric clashes (orange) aromatic-aromatic and sulfur-lone pair (both purple).
Figure 11. Comparing equilibrated 4PEP (left) and #20 (right) from the Flare FEP result. Shown here are the interactions: hydrogen bonds (green), steric clashes (orange) aromatic-aromatic and sulfur-lone pair (both purple).
Figure 11 allows us to compare the equilibrated 4PEP complex (left) with the lead #20 (right) from the Flare FEP results. Examining the interactions: the common azaindole core binds to the kinase hinge via a hydrogen bond donor to the carbonyl and an acceptor to the amine of a CYS residue. We can hypothesize from Figure 9 that adding the (magic?) Cl helps exclude water from a hydrophobic region and facilitates the lone pair on the N of the azaindole to form a sulfur-lone pair interaction with the CYS residue (circled in orange, present in #20 but not in 4PEP).
In #20 there is an overall more favourable orientation of the azaindole with respect to the above PHE and a coordination of the protonated tetrahydropyridine with water, avoiding a clash with the below ASP (circled in green, the water is present in #20 but not 4PEP). These enhancements combined with the 56o torsional twist maintained on the ortho-methoxy in #20 which maintains van der Waal contacts (this is observed to be important in CDK2 binding, pdb code 7M2F2) can likely account for the high predicted potency.
Conclusions
In this case study we show how starting with just one reference molecule with known binding and one known active as a starter for Hit Expander, you can quickly turn around a drug discovery problem from many suggestions to a few predictions with the high accuracy that can be assured when using alchemical methods such as Flare FEP.
Upon examining the discovery of potential leads, originating from one hit, whittling down from 84, we re-discover some ligands from the Tong et al. set that exhibit known (good) activity. More than that, we find results that have potential for even better activity in terms of binding CDK9 and whole cell activity and may have promise as novel drugs. ‘4PEP variant #20’ seems particularly promising, but there are 7 other reliable possibilities that could warrant further investigation.
References
- United States Patent Tong et al US9796708 Oct. 24, 2017 https://patents.google.com/patent/US9796708
- Tong et al. Balancing Properties with Carboxylates: A Lead Optimization Campaign for Selective and Orally Active CDK9 Inhibitors, ACS Med. Chem. Lett. 2021, 12, 1108-1115 doi: https://doi.org/10.1021/acsmedchemlett.1c00161
- Wang, S. and Fischer, PM. Cyclin-dependent kinase 9: a key transcriptional regulator and potential drug target in oncology, virology and cardiology. Trends in Pharmacological Sciences. 2008, 29:6, 302-313. doi: https://doi.org/10.3389/fonc.2021.678559
- Alsfouk, A. Small molecule inhibitors of cyclin-dependent kinase 9 for cancer therapy, Journal of Enzyme Inhibition and Medicinal Chemistry. 2021, 36:1, 693-706 doi: 10.1080/14756366.2021.1890726
- Nomura, T., Sumi, E., Egawa, G. et al. The efficacy of a cyclin dependent kinase 9 (CDK9) inhibitor, FIT039, on verruca vulgaris: study protocol for a randomized controlled trial. Trials 20, 489 (2019). doi: https://doi.org/10.1186/s13063-019-3570-6
- Anshabo, AT. Milne, R. Wang. S. and Albrecht, H. CDK9: A Comprehensive Review of its Biology, and its Role as a Potential Target for Anti-Cancer Agents. Frontiers in Oncology. 2021, 11, 678559, 1-24. doi: https://doi.org/10.3389/fonc.2021.678559
- Hellvard, A., Zeitlmann, L., Heiser, U. et al. Inhibition of CDK9 as a therapeutic strategy for inflammatory arthritis. Sci Rep 6, 31441 (2016). https://doi.org/10.1038/srep31441
- Vinter, J. Comput.-Aided Mol. Des., 1994, 8, 653-668
- Hu, Y.; Mügge, I. In Silico Positional Analogue Scanning with Amber GPU-TI,. J. Chem. Inf. Model. 2022, 62, 18, 4448–4459 doi: https://pubs.acs.org/doi/pdf/10.1021/acs.jcim.2c00860
- Qui Y. et al. Development and Benchmarking of Open Force Field v1.0.0-the Parsley Small-Molecule Force Field. Journal of Chemical Theory and Computation 2021, 17 (10), 6262-6280. doi: https://doi.org/10.1021/acs.jctc.1c00571
- Wagner J. et al. openforcefield/openff-forcefields: Version 2.0.0 "Sage" (2.0.0). 2021. doi: https://doi.org/10.5281/zenodo.5214478
- C. Bannwarth, E. Caldeweyher, S. Ehlert, A. Hansen, P. Pracht, J. Seibert, S. Spicher, S. Grimme, Extended tight‐binding quantum chemistry methods, Wiley Interdisciplinary Reviews: Computational Molecular Science 2021, 11, 2, e1493.
- C. Bannwarth, S. Ehlert, S. Grimme, GFN2-xTB—An accurate and broadly parametrized self-consistent tight-binding quantum chemical method with multipole electrostatics and density-dependent dispersion contributions, J. Chem. Theory Comp. 2019, 15, 3, 1652-1671 doi: https://doi.org/10.1021/acs.jctc.8b01176
- J. A. Maier, C. Martinzes, K. Kasavajhala, L. Wickstrom, K. E. Hauser, C. Simmerling, ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB, J. Chem. Theory Comput., 2015, 11, 8, 3696-3713 doi: https://doi.org/10.1021/acs.jctc.5b00255
- Liu, S., Wu, Y., Lin, T. et al. Lead optimization mapper: automating free energy calculations for lead optimization. J Comput Aided Mol Des 27, 755–770 (2013). doi: https://doi.org/10.1007/s10822-013-9678-y