flare™
Inform and guide new molecule design with electrostatics
Visual feedback on new molecule designs to understand ligand binding, structure-activity relationships and rank new molecule designs
Flare uses the XED force field to calculate a detailed map of the electrostatic character of the protein active site and your ligand. The interaction potentials provide you with vital knowledge of the fundamental processes that underlie ligand-protein binding, helping you to perfect the design of new molecules.
Electrostatic Complementarity™
- Produce surface images of proteins and ligands and calculate Electrostatic Complementarity (EC) scores and maps which can rapidly suggest activity predictions
- Quickly assess visually and numerically the EC between a protein binding site and ligand
- Improve molecule prediction and design by having visibility of the correlation between EC scoring and bioactivity for electrostatically driven interactions
- Computationally inexpensive and easy to assess large ligand data sets in a protein family to search for comparisons, trends and possible improvements
- Electrostatic Complementarity as a Fast and Effective Tool to Optimize Binding and Selectivity of Protein–Ligand Complexes J. Med. Chem. 2019, 62, 6, 3036-3050
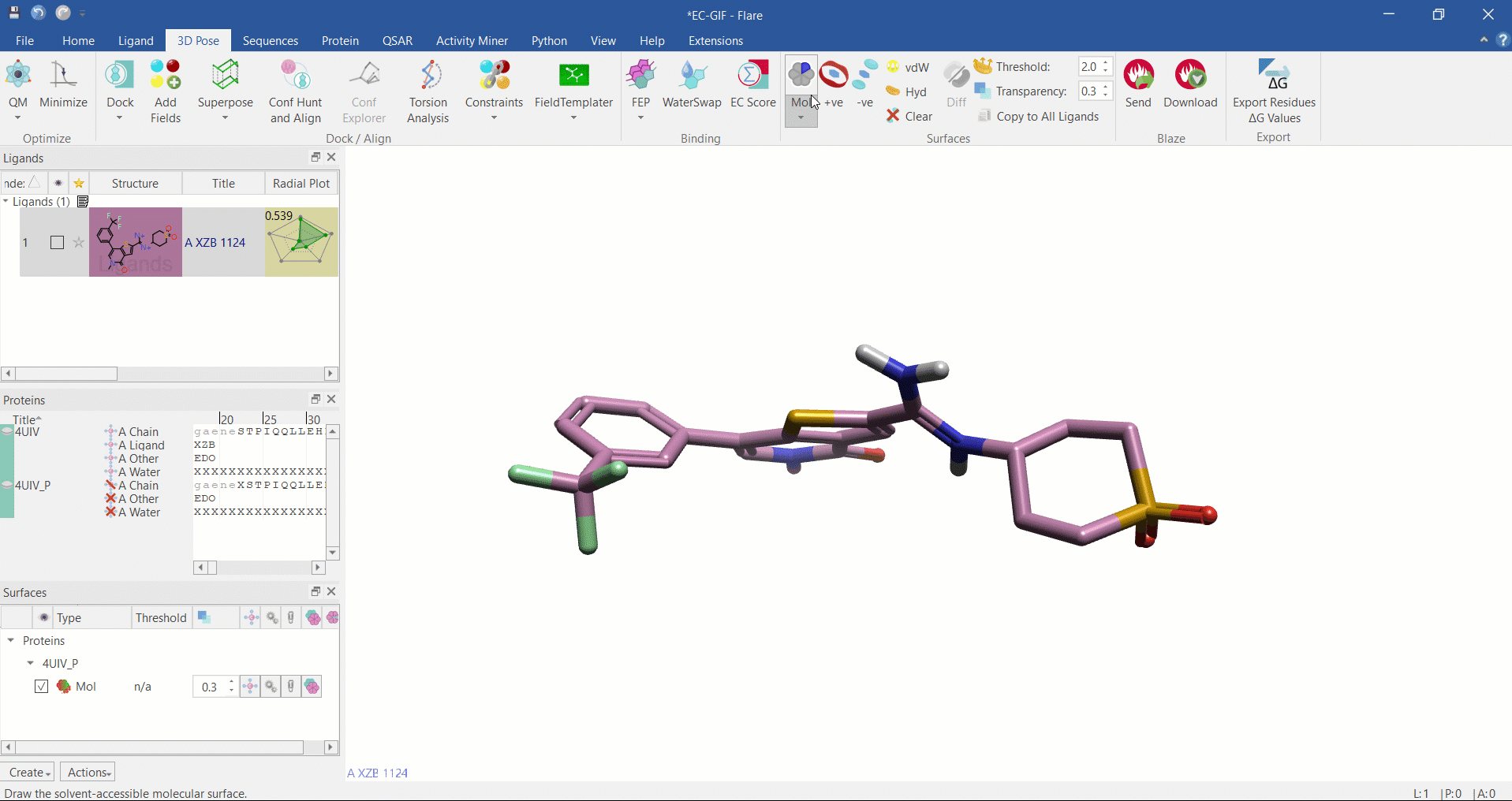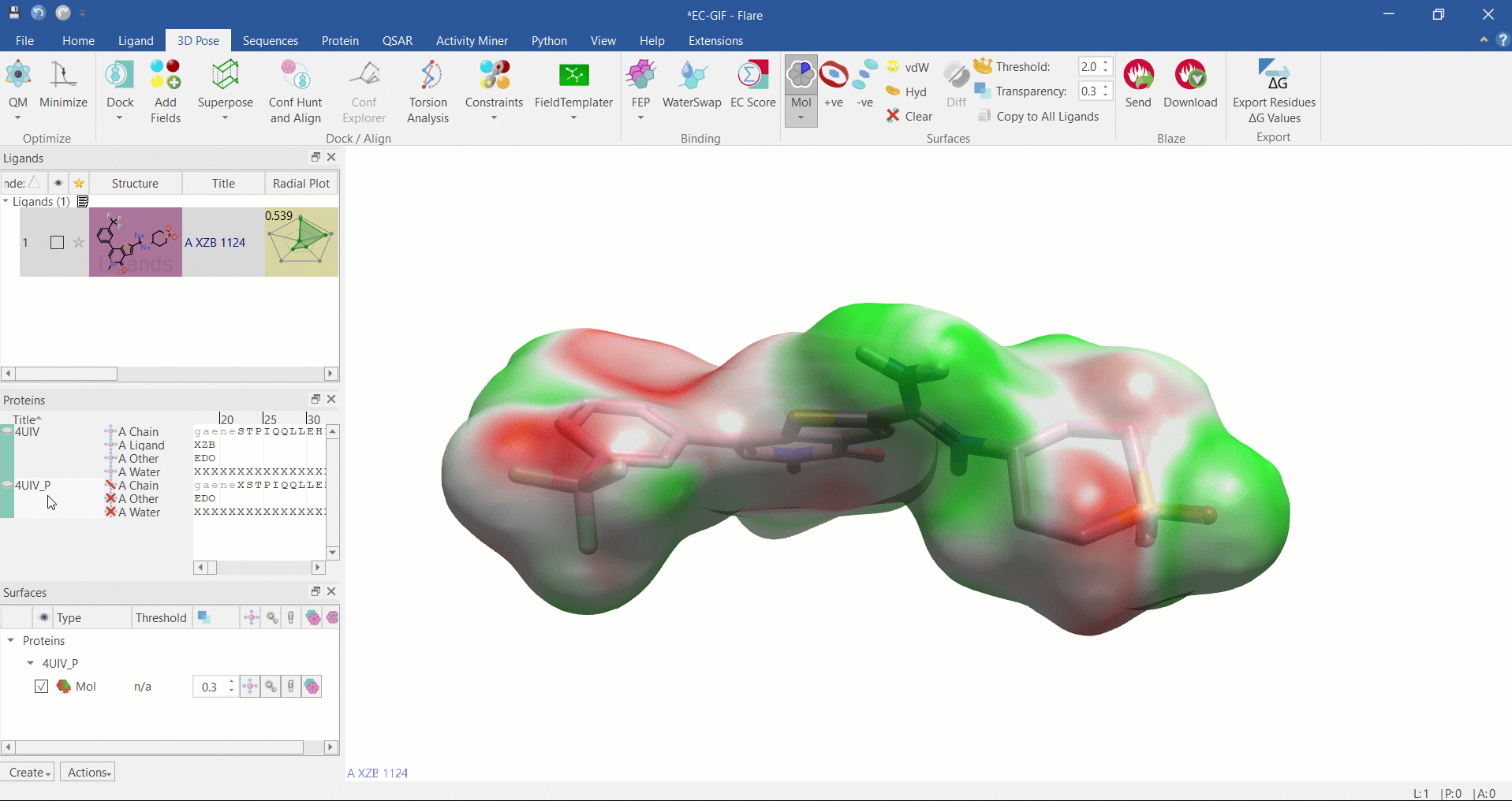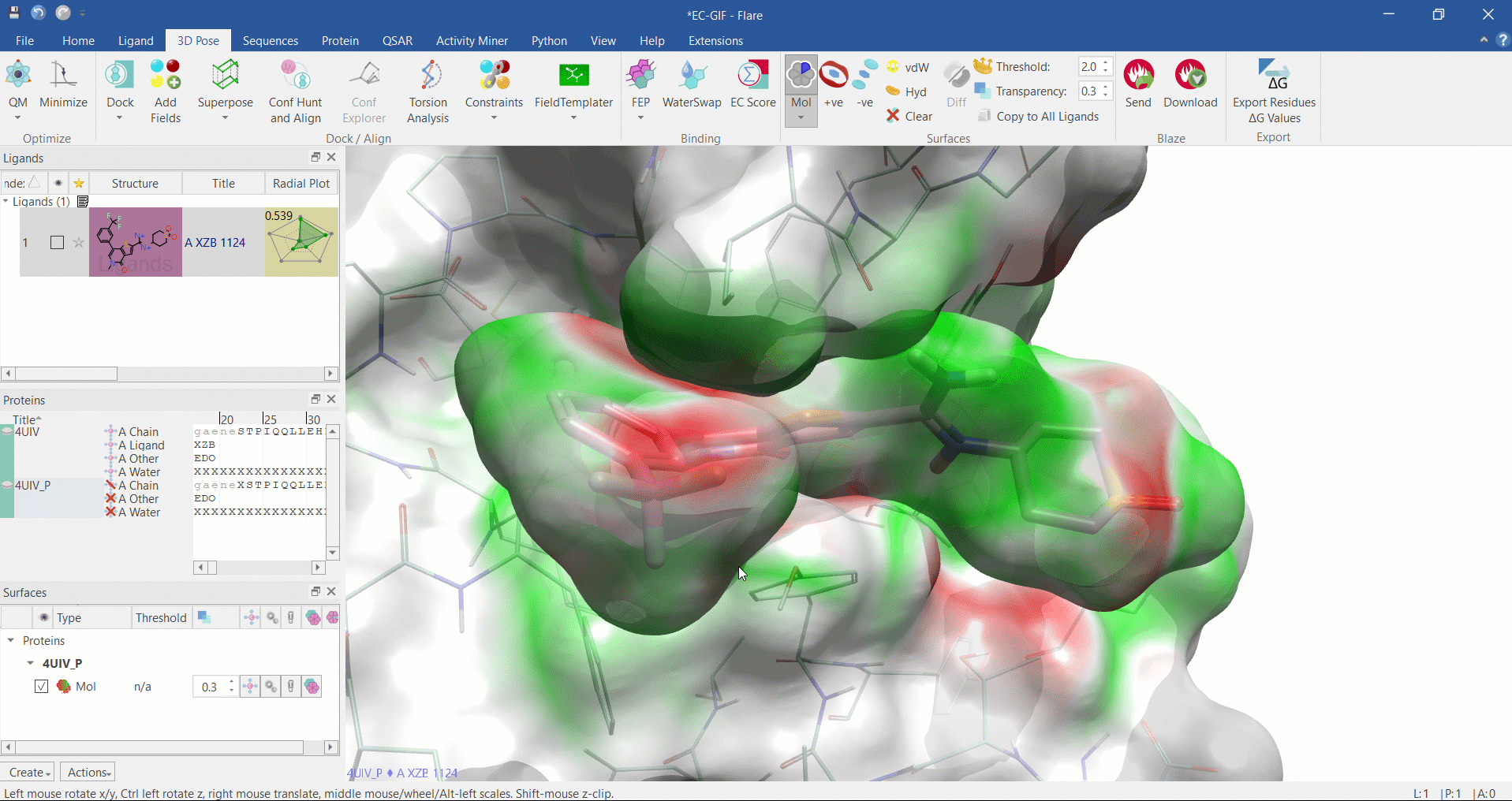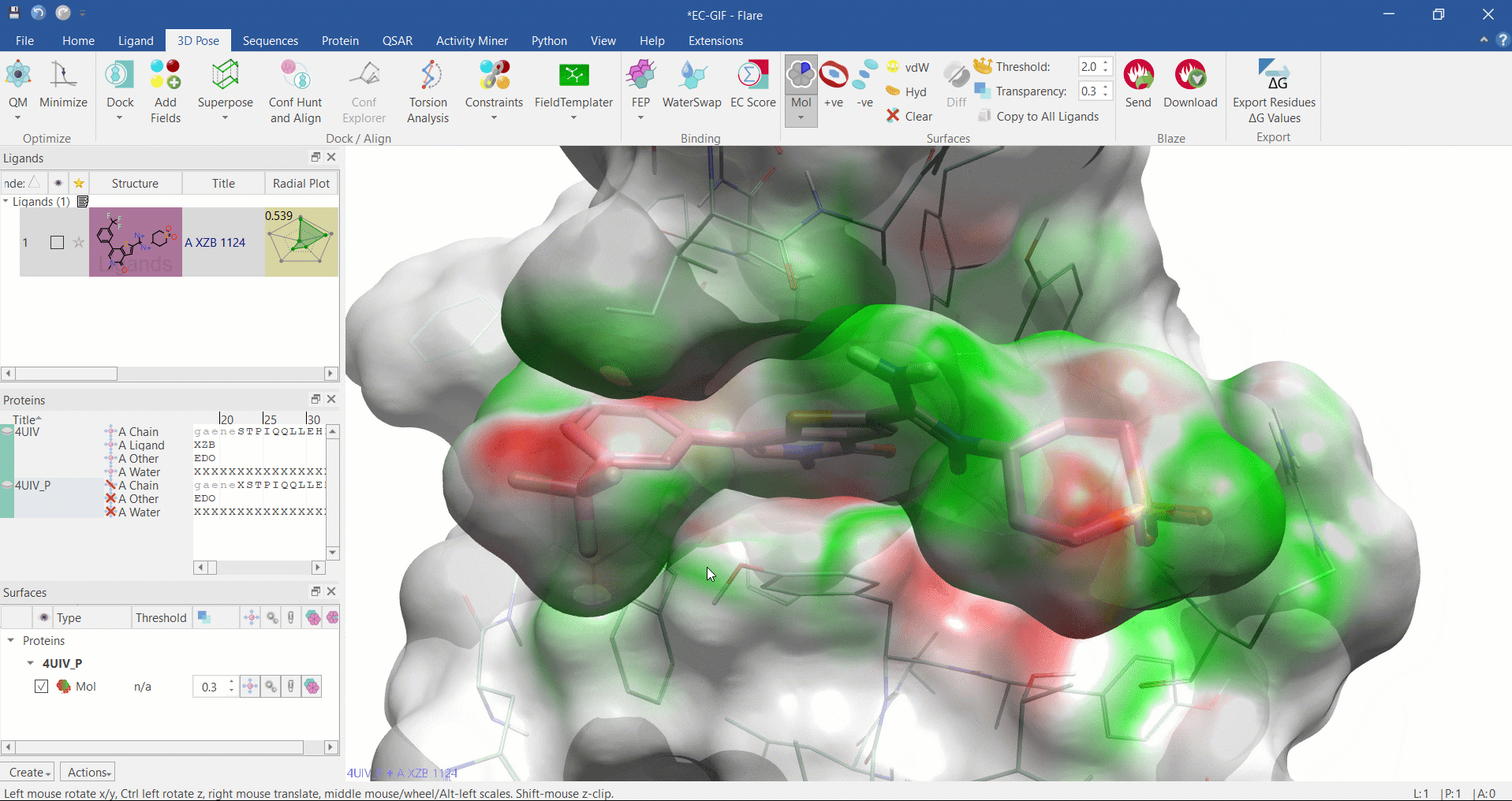
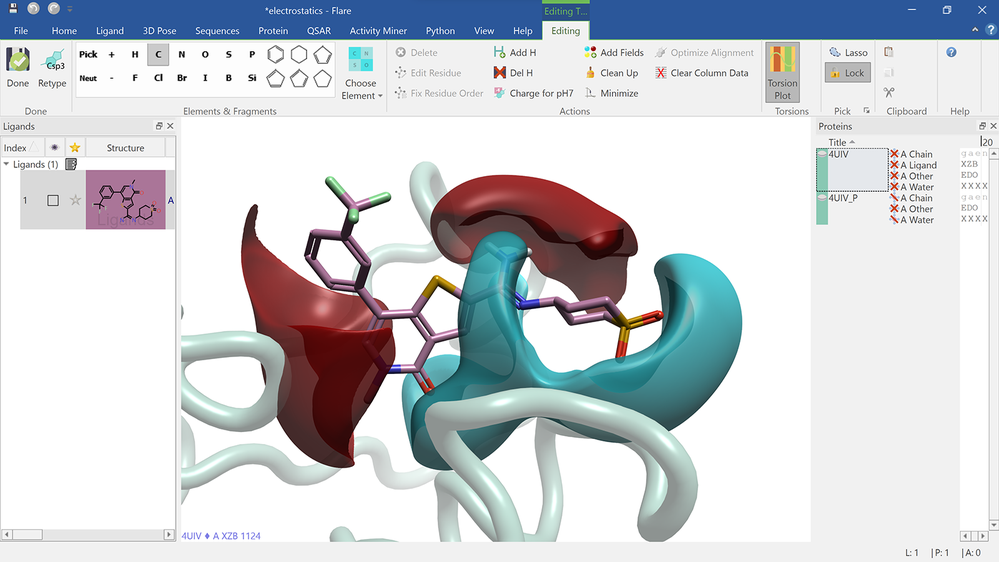
Electrostatic Complementarity surface shown on ligand and 4UIV protein
Protein interaction potentials
- Also known as ‘protein fields’, Protein interaction potentials (PIPs) are an extension of the Cresset molecular interaction potentials and map relevant ligand protein interactions
- Determine where to put ligand atoms and identify electrostatically favorable water locations
- Make predictions of selectivity, inspect mutant analysis, aid selection of protonation and tautomeric states, and optimize binding interactions by using these surface analysis tools
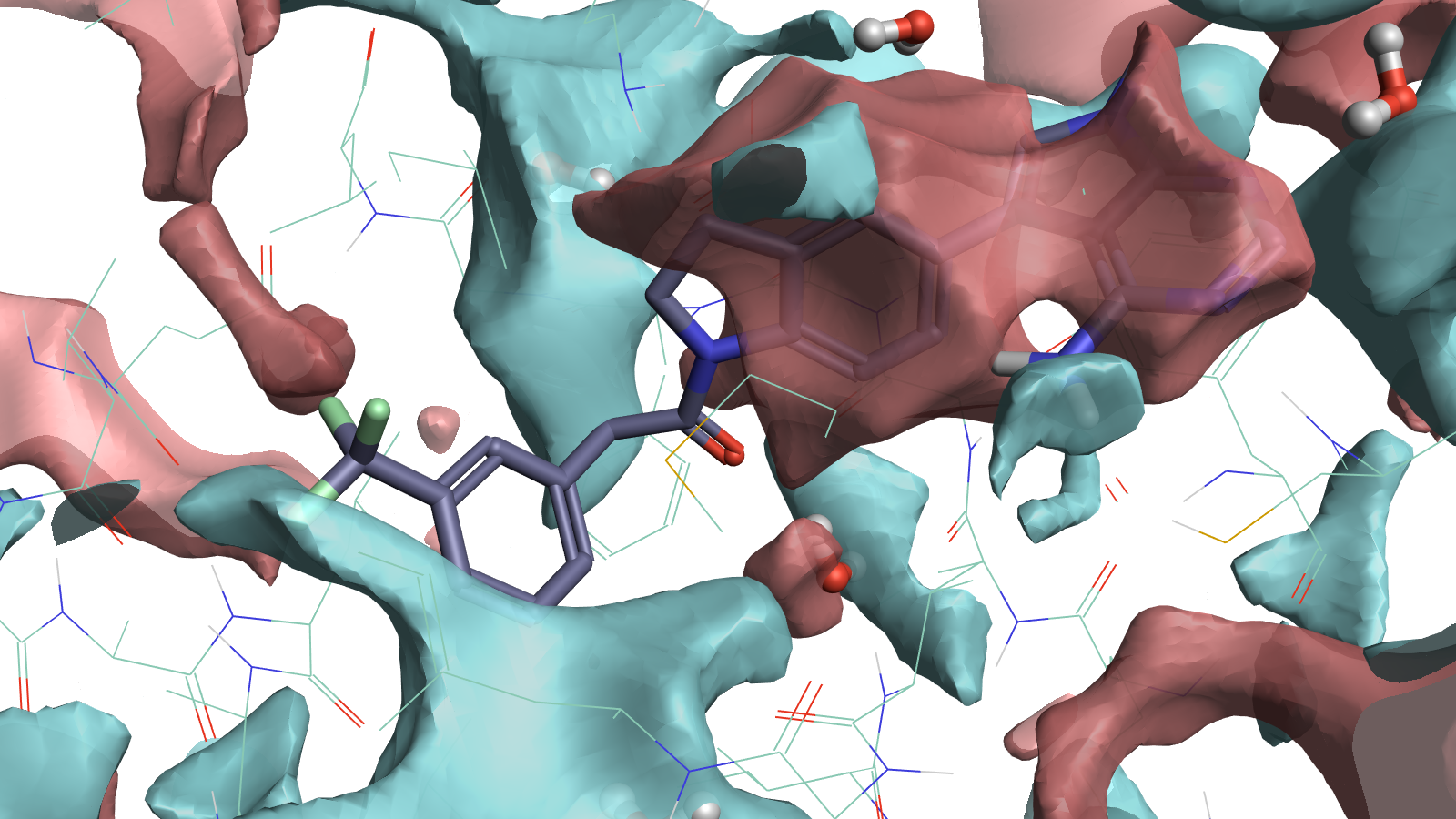
PDB entry 4G31 illustrating that full protein interaction potential maps can guide the placement of ligand atoms. Positive (red) protein surfaces map where negative (blue) ligand surfaces could fill, indicating what ligand electrostatics could work well. Electrostatically favorable water positions are also visible indicating where you might replace the water with polar interactions on the ligand.
Electrostatic Potential (ESP) surfaces
- Interaction fields can be mapped onto protein surfaces allowing quick and easy visual assessment of electrostatic trends across protein families
- Identify selectivity opportunities by comparing related proteins, using the proteins electrostatics to understand the ligand SAR better
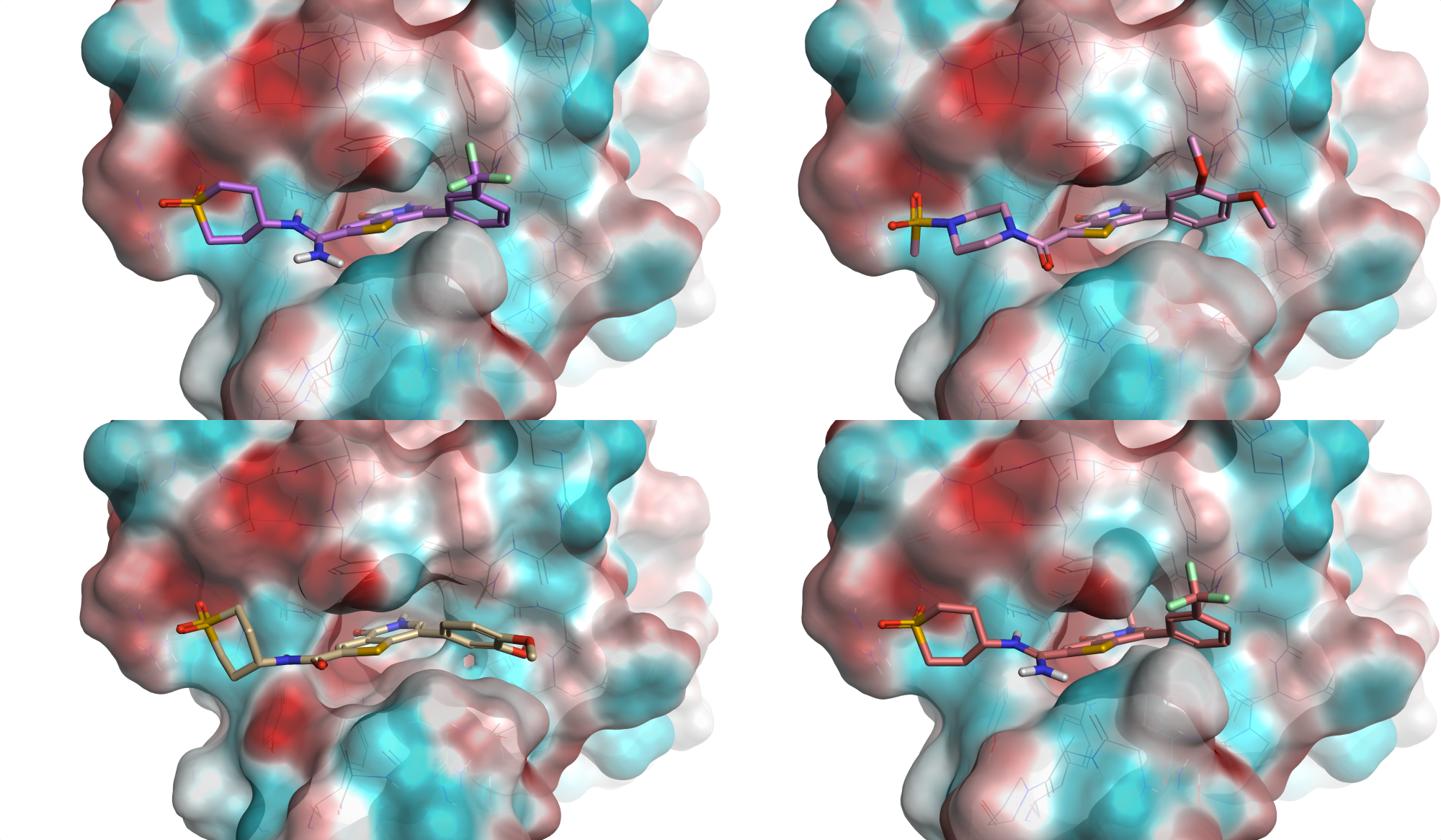
Edit new molecules informed by the electrostatic changes
Designing new ligands in Flare gives you immediate feedback on electrostatic changes while viewing your molecule in the protein active site.