software
Flare™ Free Energy Perturbation (FEP)
Use Flare FEP to make the right ligand design choices and enable lead optimization with confidence
A robust and accurate method for calculating binding affinity
Flare Free Energy Perturbation (FEP) offers a quantitative method to reliably calculate relative binding affinity, enabling accurate ranking of molecules in a congeneric ligand series. The method enables users to test 'in-silico' a large number of molecules, prior to focusing on ‘wet’ lab work, meaning that fewer compounds need to be made and tested to achieve the desired results.
The process supports chemists in making known actives more potent, without having to synthesize hundreds or thousands of compounds, eliminating the time wasted on synthesizing non-potent molecules. Benchmarking FEP experiments can be used to gain confidence that your system is prepared correctly, confirming the predictivity of the method on the target and ligand series of interest. Then production FEP experiments yield predicted binding free energies for new molecular designs.
FEP calculation results typically fall within 1kcal/ mol from experimental data. This means that Flare users can make better, accurately informed decisions around which ligand modifications can achieve the best results, with new molecule designs often significantly different from the initial molecular structure and with enhanced potency.
- Reduce the amount of ‘wet’ chemistry across all stages of drug discovery
- Help prioritize compounds for synthesis and greatly reduce the time and cost involved in a drug discovery project
- Increase the efficiency of designing new compounds for key therapeutic targets
- Accurately predict ligand binding affinity
- Find drug candidates with enhanced potency at the target of interest
- Effectively triage and prioritize the most promising candidates for further investigation, thanks to Flare FEP’s integration with other software solutions and modules such as Hit Expander and Spark™
Flare licensing options
Robust and efficient calculations
- Flare’s accessible, user-friendly interface enables chemists to take full advantage of free energy calculations, regardless of their technical background
- Seamlessly prepare proteins and ligands for the FEP experiment
- Easily generate robust ligand poses with the ligand alignment component of Flare, or by docking ligands to the protein you will use in the FEP experiment
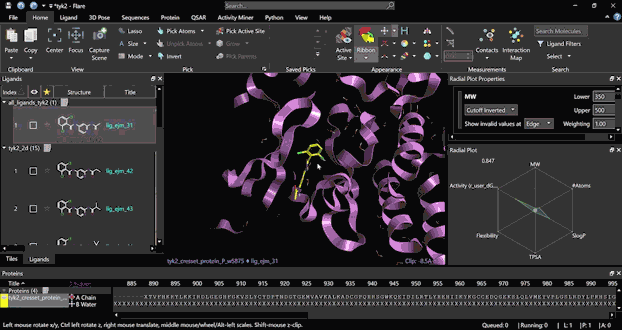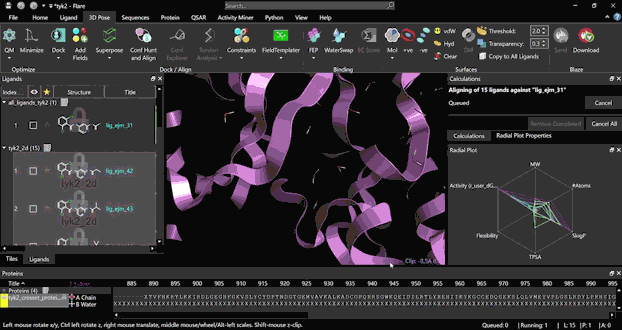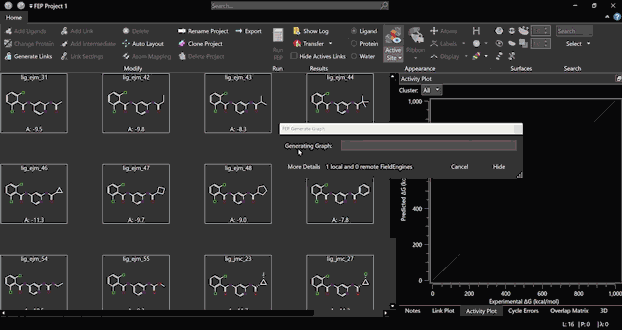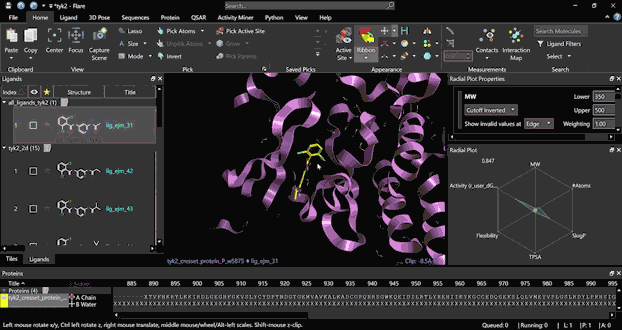
- Use Flare FEP’s automated algorithms to create an efficient perturbation network quickly, generating intermediate ligands when needed
- Multiple options to refine and optimize the perturbation network
- Easily view and troubleshoot all aspects of your Flare FEP project with dedicated, highly visual tools
- View and prioritize your ligands based on their predicted activity
Run an FEP experiment starting from scratch with a few simple steps, thanks to the platform’s user-friendly interface
Accurate FEP calculations
- Accurately calculate the relative binding free energy of a congeneric series of ligands (Assessment of Binding Affinity via Alchemical Free-Energy Calculations J. Chem. Inf. Model. 2020, 60, 6, 3120–3130)
- Assess the accuracy of your FEP model by comparing experimental versus predicted activity values using Flare FEP’s ‘activity plot’
- Precision is obtained in your Flare FEP calculations using fully connected, two-way networks, which enable more accurate error estimation for each link and thorough analysis of cycle errors and hysteresis
- Enhance predictive performance by improving the water sampling of occluded pockets in the protein active site using Grand Canonical Nonequilibrium Candidate Monte Carlo (GCNCMC)
- Automatically create custom torsion parameters for small molecules using GFN2-xTB extended semiempirical tight-binding, ANI-2x deep learning QM approximation or hybrid DFT//GFN2-xTB calculations
- Facilitate a challenging transformation by automatically adding intermediate molecules to better link structurally related ligands
- 3D visualization tools allow for close inspection of the initial and final states of the ligands to ensure that no unrealistic or undesirable geometries are adopted
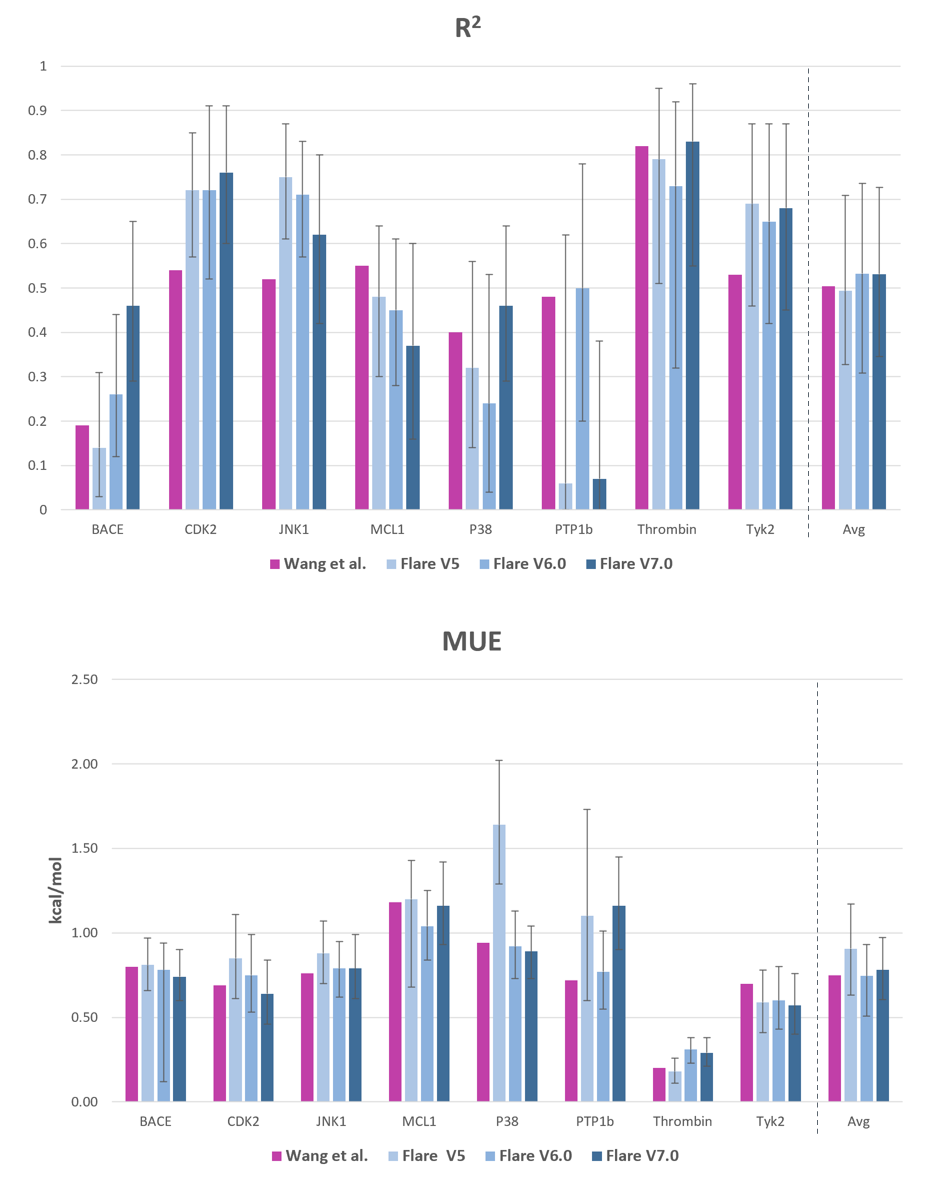
Comparison of Flare FEP's accuracy in relation to previous versions of Flare (depicted in shades of blue) and the results of Wang et al. (illustrated in purple)
Achieving a balance between technical sophistication, software usability and computational cost
- Harness the power of Graphical Processing Units (GPUs) or cloud-based computing resources such as Amazon Web Services (AWS)
- Optimize calculation time by using the Cresset Engine Broker™ to connect to a local cluster or cloud computing facilities
- Keep computational costs to a minimum, thanks to the ‘adaptive lambda window’ algorithm which ensures the minimum number of lambda windows is used for each calculation, without affecting accuracy
Contact us to request a free evaluation of Flare FEP, or to learn more about our licensing options
References and acknowledgements
P. Eastman, J. Swails, J. D. Chodera, R. T. McGibbon, Y. Zhao, K. A. Beauchamp, L.-P. Wang, A. C. Simmonett, M. P. Harrigan, C. D. Stern, R. P. Wiewiora, B. R. Brooks, and V. S. Pande, OpenMM 7: Rapid development of high performance algorithms for molecular dynamics, PLOS Comp. Biol. 2017, 3(7): e1005659
S. Liu, Y. Wu, T. Lin, R. Abel, J. P. Redmann, C. M. Summa, V. R. Jaber, N. M. Lim, D. L. Mobley, Lead optimization mapper: automating free energy calculations for lead optimization, J. Comput. Aided Mol. Des. 2013, 27, 9, 755-770
L. O. Hedges, A. S. J. S Mey, , C. A. Laughton, F. L. Gervasio, A. J. Mulholland,C. J. Woods, J. Michel, BioSimSpace: An interoperable Python framework for biomolecular simulation, Journal of Open Source Software 2019, 4(43), 1831
C. Woods , L. O. Hedges, A. S. J. S. Mey , G. Calabrò and J. Michel , www.siremol.org, Sire Molecular Simulation Framework , 2022
G. Calabrò , C. J. Woods , F. Powlesland , A. S. J. S. Mey , A. J. Mulholland and J. Michel , Elucidation of Nonadditive Effects in Protein–Ligand Binding Energies: Thrombin as a Case Study, J. Phys. Chem. B 2016, 120 , 5340 —5350
H. H. Loeffler, J. Michel, C. J. Woods, FESetup: Automating Setup for Alchemical Free Energy Simulations, J. Chem. Inf Model. 2015, 55 (12), 2485-2490
M. L. Samways, H. E. Bruce Macdonald, J. W. Essex, grand: A Python Module for Grand Canonical Water Sampling in OpenMM, J. Chem. Inf. Model. 2020, 60, 10, 4436-4441
O. J. Melling, M. L. Samways, Y. Ge, D. L. Mobley, J. W. Essex, Enhanced Grand Canonical Sampling of Occluded Water Sites Using Nonequilibrium Candidate Monte Carlo, J. Chem. Theory Comput. 2023, 19, 1050-1062
C. Bannwarth, S. Ehlert, S. Grimme, GFN2-xTB—An accurate and broadly parametrized self-consistent tight-binding quantum chemical method with multipole electrostatics and density-dependent dispersion contributions, J. Chem. Theory Comp. 2019, 15, 3, 1652-1671
C. Devereux, J. S. Smith, K. K. Huddleston, K. Barros, R. Zubatyuk, O. Isayev, and A. E. Roitberg, Extending the applicability of the ANI deep learning molecular potential to sulfur and halogens, J. Chem. Theory Comput. 2020, 16,7, 4192–4202
P. K. Behara, H. Jang, J. Horton, D. Dotson, S. Boothroyd, C. Cavender, V. Gapsys, T. Gokey, D. Hahn, J. Maat, O. Madin, I. Pulido, M. Thompson, J. Wagner, L. Wang, J. Chodera, D. Cole, M. Gilson, M. Shirts, C. Bayly, L.-P. Wang, D. Mobley, Benchmarking QM theory for drug-like molecules to train force fields, Poster presented at CUP XXI, March 08, 2022 - March 10, 2022, Santa Fe, New Mexico, USA
L. Wang, Y. Wu, Y. Deng, B. Kim, L. Pierce, G. Krilov, D. Lupyan, S. Robinson, M. K. Dahlgren, J. Greenwood, D. L. Romero, C. Masse, J. L. Knight, T. Steinbrecher, T. Beuming, W. Damm, E. Harder, W. Sherman, M. Brewer, R. Wester, M. Murcko, L. Frye, R. Farid, T. Lin, D. L. Mobley, W. L. Jorgensen, B. J. Berne, R. A. Friesner, R. Abel, Accurate and Reliable Prediction of Relative Ligand Binding Potency in Prospective Drug Discovery by Way of a Modern Free-Energy Calculation Protocol and Force Field, J. Am. Chem. Soc. 2015, 137, 2695−2703