flare™
Find druggable binding sites in your protein
Use pocket detection and analysis in Flare to identify druggable binding sites in your protein to exploit in your drug discovery strategy
The identification and characterization of potential druggable binding sites for small molecule ligands is a key step in the molecular modeling of novel targets and lead series.
Pocket detection and analysis in Flare uses fpocket, a fast protein pocket detection algorithm for the identification and characterization of pockets and cavities within a protein structure. It can be run on individual protein structures experimentally derived by X-ray, modeled by homology, or obtained as a snapshot from molecular dynamics studies.
Using the mdpocket extension of the method, pocket analysis can be run or on conformation ensembles from Molecular Dynamics, to monitor the frequency of opening/closing and the druggability of pockets.
- Find the druggable binding sites in your protein
- Search for different types of pockets, including drug binding sites, water binding pockets, channels and small cavities and large solvent exposed sites
- Characterize each pocket according to different parameters, including a druggability score
- Monitor the frequency of opening/closing of druggable binding pockets over a Molecular Dynamics trajectory
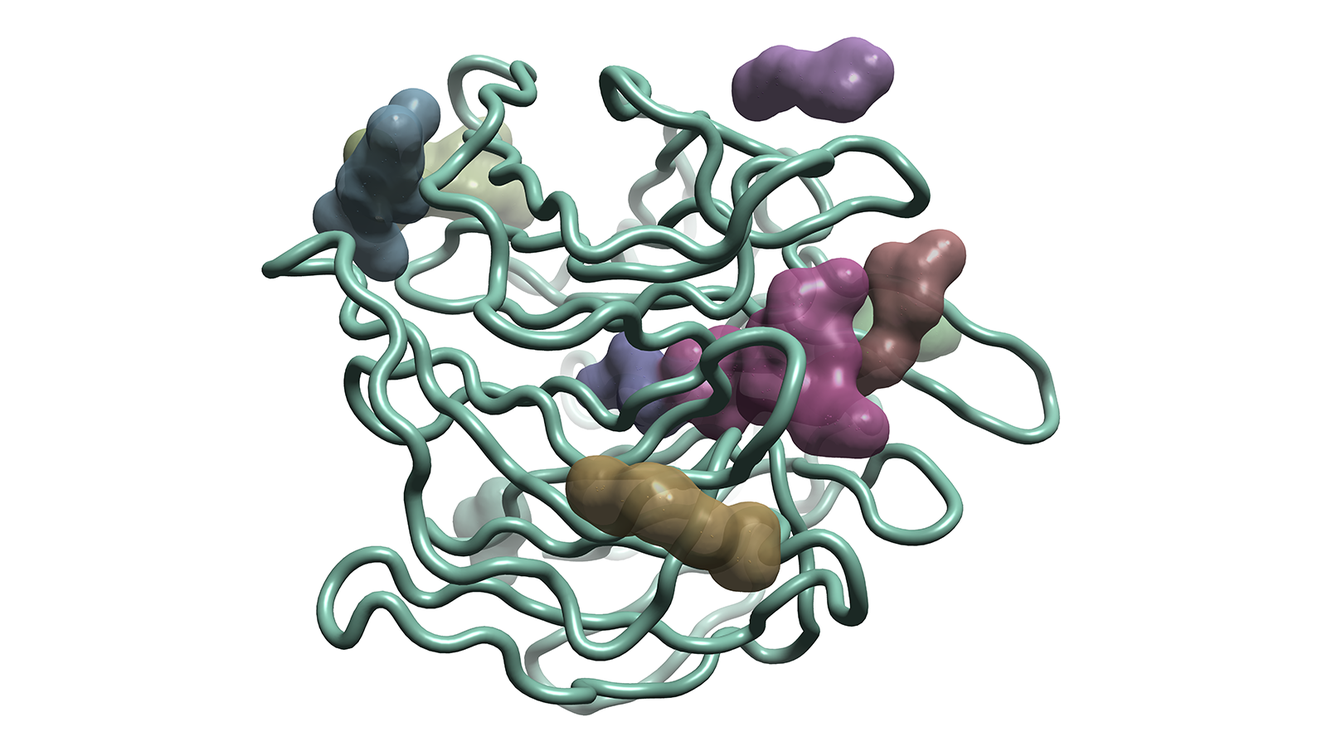
Search for pockets or cavities in individual protein structures, including drug binding sites, water binding pockets, channels and large solvent-exposed sites
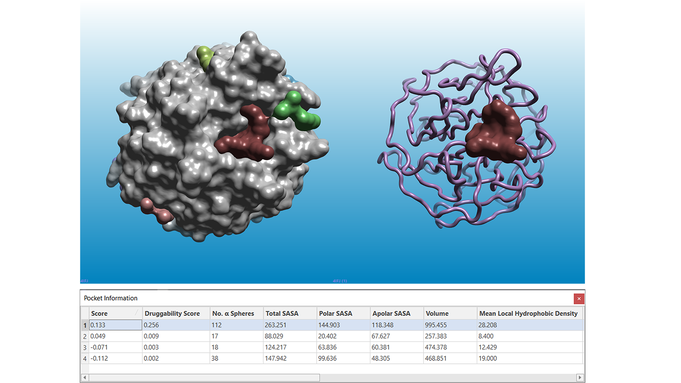
Results of a pocket detection experiment ran on the apo structure of KEAP1 (PDB: 4IFJ). Left: the solvent excluded surface of the KEAP1 protein is shown in grey. Right: the KEAP1 protein structure is shown as a pink ribbon. In both pictures, pockets are shown as multi-color surfaces. The pocket information table (bottom) reports the characterization of the pockets according to a number of different parameters.
References and acknowledgements
V. Le Guilloux, P. Schmidtke and P. Tuffery, Fpocket: An open source platform for ligand pocket detection BMC Bioinformatics 2009, 10, 168
P. Schmldtke, A. Bidon-Chanal, J. Luque, X. Barril, MDpocket: open-source cavity detection and characterization on molecular dynamics trajectories Bioinformatics. 2011, 27, 3276-85