Cresset software solutions are available through easy to use graphical user interfaces, from the command line and through workflow solutions such as KNIME™. Using the KNIME interface enables integration of our approach into standard workflows and with other applications. We provide access to our applications through KNIME nodes that call the corresponding command line application behind the scenes.
Spark™ nodes
Spark enables you to generate highly innovative ideas for your project to explore chemical space and escape IP and toxicity traps.
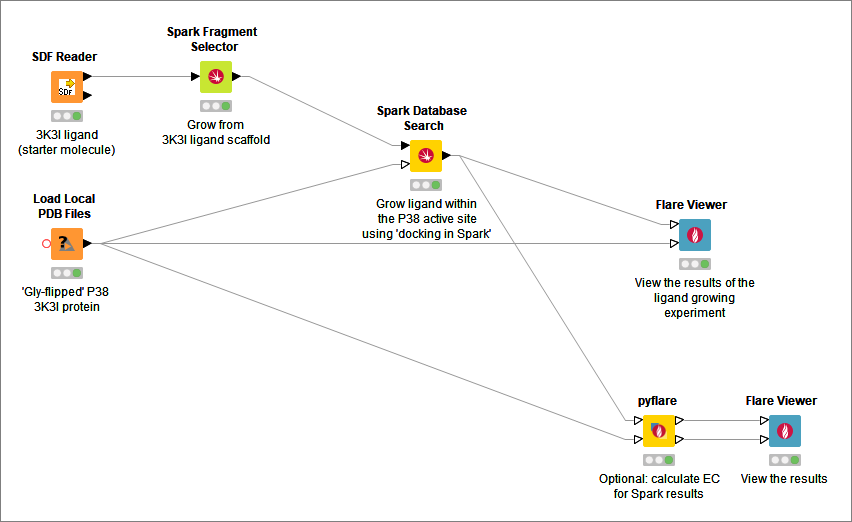
Given a ‘starter molecule’, you can select a portion to replace and specify which databases to search. Spark will suggest a list of new molecules containing replacement moieties with similar electrostatic and steric properties ranked using Cresset’s unique molecular field technology, or picking new ligand-protein interactions from the active site using the docking in Spark feature. In addition you can generate your own databases of replacement moieties using the database generator functionality.
In KNIME the Spark nodes provide access to the scaffold hopping and R-group exploring capabilities with a specialized node to enable graphical specification of the portion of the starter molecule to be replaced. A database creation node is also included.
- Spark Database Search: search databases for novel scaffolds or R-groups for a specified molecule
- Spark Fragment Selector: enables the graphical selection of the region to be replaced by Spark in a database search experiment
- Generate Spark Database: generate Spark databases of fragments or reagents
Flare™ nodes
Flare is an integrated platform for ligand-based and structure-based drug design that enables research chemists to discover novel small molecules more efficiently and effectively.
In KNIME the Flare nodes enable ligand comparisons and investigation, building of qualitative and quantitative 3D models of activity, scoring of new molecule designs, docking and scoring experiments, and enable full access to the Flare functionality through the Flare Python API.
- Flare Align: Align and score molecules to a 3D reference molecule using electrostatics and shape or common substructure. Molecules are conformationally hunted if necessary
- Flare Docking: run docking and scoring experiments using the Lead Finder™ docking algorithm
- Flare Build/Score Field QSAR: generate 3D-QSAR models, and predict activities for new molecules using a Flare 3D-QSAR model
- Flare Build/Score kNN: generate kNN regression or classification models, and predict activities for new molecules using a Flare kNN model
- Flare Build/Score Activity Atlas™: generate qualitative Activity Atlas models, and predict the novelty of new molecules using an Activity Atlas model
- Flare Field Descriptors: generate Cresset 3D field descriptors for aligned ligands which can be used to build machine learning models
- pyflare: Run a Python script which has access the Flare Python API
The full list of Flare nodes can be found in the release notes.
XedTools™ nodes
The XedTools nodes give access to core technologies such as ligand minimization and conformation generation.
- XedMin™ is a ligand minimizer (using Cresset’s proprietary XED force field) to optimize ligands in isolation or in a specified context such as the protein active site
- XedeX™ creates low energy, diverse conformation populations for a ligand
Viewer nodes
Cresset supplies a generic Flare Viewer node that calls the main Flare graphical user interface on the client machine. This node is useful for visualizing ligand and protein structures or for viewing the results of other Cresset nodes.
Installation
See KNIME node installation instructions or refer to the latest release notes.
Licensing
The KNIME nodes require you to accept the terms of our license agreement when they are installed but do not require further license files. However, the software used by the KNIME nodes does require a valid license file which will be supplied by Cresset. If you are a customer and cannot locate your license file, then please contact us. Alternatively, request an evaluation license.
Examples
Download example workflows that encompass each of the Cresset KNIME nodes.